SPA: Short peptide analyzer of intrinsic disorder status of short peptides
- PMID: 20497238
- PMCID: PMC2900848
- DOI: 10.1111/j.1365-2443.2010.01407.x
SPA: Short peptide analyzer of intrinsic disorder status of short peptides
Abstract
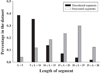
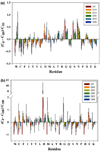
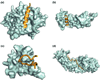
Disorder prediction for short peptides is important and difficult. All modern predictors have to be optimized on a preselected dataset prior to prediction. In the succeeding prediction process, the predictor works on a query sequence or its short segment. For implementing the prediction smoothly and obtaining sound prediction results, a specific length of the sequence or segment is usually required. The need of the preselected dataset in the optimization process and the length limitation in the prediction process restrict predictors' performance. To minimize the influence of these limitations, we developed a method for the prediction of intrinsic disorder in short peptides based on large dataset sampling and statistics. As evident from the data analysis, this method provides more reliable prediction of the intrinsic disorder status of short peptides.
Figures


Similar articles
-
Structural prediction of peptides binding to MHC class I molecules.Proteins. 2006 Apr 1;63(1):43-52. doi: 10.1002/prot.20870. Proteins. 2006. PMID: 16447245
-
Improving Sequence-Based Prediction of Protein-Peptide Binding Residues by Introducing Intrinsic Disorder and a Consensus Method.J Chem Inf Model. 2018 Jul 23;58(7):1459-1468. doi: 10.1021/acs.jcim.8b00019. Epub 2018 Jun 25. J Chem Inf Model. 2018. PMID: 29895149
-
Evaluation of MHC-II peptide binding prediction servers: applications for vaccine research.BMC Bioinformatics. 2008 Dec 12;9 Suppl 12(Suppl 12):S22. doi: 10.1186/1471-2105-9-S12-S22. BMC Bioinformatics. 2008. PMID: 19091022 Free PMC article.
-
Atomic displacement parameters in structural biology.Amino Acids. 2018 Jul;50(7):775-786. doi: 10.1007/s00726-018-2574-y. Epub 2018 May 11. Amino Acids. 2018. PMID: 29752562 Review.
-
Uncovering new aspects of protein interactions through analysis of specificity landscapes in peptide recognition domains.FEBS Lett. 2012 Aug 14;586(17):2764-72. doi: 10.1016/j.febslet.2012.03.054. Epub 2012 Apr 3. FEBS Lett. 2012. PMID: 22710167 Review.
Cited by
-
An Overview of Predictors for Intrinsically Disordered Proteins over 2010-2014.Int J Mol Sci. 2015 Sep 29;16(10):23446-62. doi: 10.3390/ijms161023446. Int J Mol Sci. 2015. PMID: 26426014 Free PMC article. Review.
-
Effective identification of Gram-negative bacterial type III secreted effectors using position-specific residue conservation profiles.PLoS One. 2013 Dec 31;8(12):e84439. doi: 10.1371/journal.pone.0084439. eCollection 2013. PLoS One. 2013. PMID: 24391954 Free PMC article.
References
-
- Balla S, Thapar V, Verma S, Luong T, Faghri T, Huang CH, Rajasekaran S, del Campo JJ, Shinn JH, Mohler WA, Maciejewski MW, Gryk MR, Piccirillo B, Schiller SR, Schiller MR. Minimotif Miner: a tool for investigating protein function. Nat. Methods. 2006;3:175–177. - PubMed
-
- Burley SK, Petsko GA. Aromatic-aromatic interaction: a mechanism of protein structure stabilization. Science. 1985;229:23–28. - PubMed
-
- Dunker AK, Brown CJ, Lawson JD, Iakoucheva LM, Obradovic Z. Intrinsic disorder and protein function. Biochemistry. 2002a;41:6573–6582. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

