Dynamic cross-talk analysis among TNF-R, TLR-4 and IL-1R signalings in TNFalpha-induced inflammatory responses
- PMID: 20497537
- PMCID: PMC2889840
- DOI: 10.1186/1755-8794-3-19
Dynamic cross-talk analysis among TNF-R, TLR-4 and IL-1R signalings in TNFalpha-induced inflammatory responses
Abstract
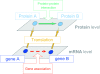
Background: Development in systems biology research has accelerated in recent years, and the reconstructions for molecular networks can provide a global view to enable in-depth investigation on numerous system properties in biology. However, we still lack a systematic approach to reconstruct the dynamic protein-protein association networks at different time stages from high-throughput data to further analyze the possible cross-talks among different signaling/regulatory pathways.
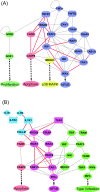
Methods: In this study we integrated protein-protein interactions from different databases to construct the rough protein-protein association networks (PPANs) during TNFalpha-induced inflammation. Next, the gene expression profiles of TNFalpha-induced HUVEC and a stochastic dynamic model were used to rebuild the significant PPANs at different time stages, reflecting the development and progression of endothelium inflammatory responses. A new cross-talk ranking method was used to evaluate the potential core elements in the related signaling pathways of toll-like receptor 4 (TLR-4) as well as receptors for tumor necrosis factor (TNF-R) and interleukin-1 (IL-1R).
Results: The highly ranked cross-talks which are functionally relevant to the TNFalpha pathway were identified. A bow-tie structure was extracted from these cross-talk pathways, suggesting the robustness of network structure, the coordination of signal transduction and feedback control for efficient inflammatory responses to different stimuli. Further, several characteristics of signal transduction and feedback control were analyzed.
Conclusions: A systematic approach based on a stochastic dynamic model is proposed to generate insight into the underlying defense mechanisms of inflammation via the construction of corresponding signaling networks upon specific stimuli. In addition, this systematic approach can be applied to other signaling networks under different conditions in different species. The algorithm and method proposed in this study could expedite prospective systems biology research when better experimental techniques for protein expression detection and microarray data with multiple sampling points become available in the future.
Figures





References
-
- Kitano H. Robustness from top to bottom. Nat Genet. 2006;38:133–133. doi: 10.1038/ng0206-133. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

