Development of genome-specific primers for homoeologous genes in allopolyploid species: the waxy and starch synthase II genes in allohexaploid wheat (Triticum aestivum L.) as examples
- PMID: 20497560
- PMCID: PMC2890506
- DOI: 10.1186/1756-0500-3-140
Development of genome-specific primers for homoeologous genes in allopolyploid species: the waxy and starch synthase II genes in allohexaploid wheat (Triticum aestivum L.) as examples
Abstract
Background: In allopolypoid crops, homoeologous genes in different genomes exhibit a very high sequence similarity, especially in the coding regions of genes. This makes it difficult to design genome-specific primers to amplify individual genes from different genomes. Development of genome-specific primers for agronomically important genes in allopolypoid crops is very important and useful not only for the study of sequence diversity and association mapping of genes in natural populations, but also for the development of gene-based functional markers for marker-assisted breeding. Here we report on a useful approach for the development of genome-specific primers in allohexaploid wheat.
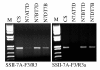
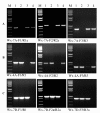
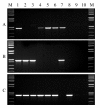
Findings: In the present study, three genome-specific primer sets for the waxy (Wx) genes and four genome-specific primer sets for the starch synthase II (SSII) genes were developed mainly from single nucleotide polymorphisms (SNPs) and/or insertions or deletions (Indels) in introns and intron-exon junctions. The size of a single PCR product ranged from 750 bp to 1657 bp. The total length of amplified PCR products by these genome-specific primer sets accounted for 72.6%-87.0% of the Wx genes and 59.5%-61.6% of the SSII genes. Five genome-specific primer sets for the Wx genes (one for Wx-7A, three for Wx-4A and one for Wx-7D) could distinguish the wild type wheat and partial waxy wheat lines. These genome-specific primer sets for the Wx and SSII genes produced amplifications in hexaploid wheat, cultivated durum wheat, and Aegilops tauschii accessions, but failed to generate amplification in the majority of wild diploid and tetraploid accessions.
Conclusions: For the first time, we report on the development of genome-specific primers from three homoeologous Wx and SSII genes covering the majority of the genes in allohexaploid wheat. These genome-specific primers are being used for the study of sequence diversity and association mapping of the three homoeologous Wx and SSII genes in natural populations of both hexaploid wheat and cultivated tetraploid wheat. The strategies used in this paper can be used to develop genome-specific primers for homoeologous genes in any allopolypoid species. They may be also suitable for (i) the development of gene-specific primers for duplicated paralogous genes in any diploid species, and (ii) the development of allele-specific primers at the same gene locus.
Figures



Similar articles
-
Molecular and phylogenetic characterization of the homoeologous EPSP Synthase genes of allohexaploid wheat, Triticum aestivum (L.).BMC Genomics. 2015 Oct 23;16:844. doi: 10.1186/s12864-015-2084-1. BMC Genomics. 2015. PMID: 26492960 Free PMC article.
-
Genome-specific primer sets for starch biosynthesis genes in wheat.Theor Appl Genet. 2004 Oct;109(6):1295-1302. doi: 10.1007/s00122-004-1743-4. Theor Appl Genet. 2004. PMID: 15340684
-
Molecular characterization of new waxy mutants identified in bread and durum wheat.Theor Appl Genet. 2005 May;110(8):1481-9. doi: 10.1007/s00122-005-1983-y. Epub 2005 Apr 16. Theor Appl Genet. 2005. PMID: 15834696
-
PCR-based landmark unique gene (PLUG) markers effectively assign homoeologous wheat genes to A, B and D genomes.BMC Genomics. 2007 May 30;8:135. doi: 10.1186/1471-2164-8-135. BMC Genomics. 2007. PMID: 17535443 Free PMC article.
-
Enrichment and Diversification of the Wheat Genome via Alien Introgression.Plants (Basel). 2024 Jan 23;13(3):339. doi: 10.3390/plants13030339. Plants (Basel). 2024. PMID: 38337872 Free PMC article. Review.
Cited by
-
Molecular and phylogenetic characterization of the homoeologous EPSP Synthase genes of allohexaploid wheat, Triticum aestivum (L.).BMC Genomics. 2015 Oct 23;16:844. doi: 10.1186/s12864-015-2084-1. BMC Genomics. 2015. PMID: 26492960 Free PMC article.
-
Homolog-specific PCR primer design for profiling splice variants.Nucleic Acids Res. 2011 May;39(10):e69. doi: 10.1093/nar/gkr127. Epub 2011 Mar 16. Nucleic Acids Res. 2011. PMID: 21415011 Free PMC article.
-
A null allele of granule bound starch synthase (Wx-B1) may be one of the major genes controlling chapatti softness.PLoS One. 2021 Jan 28;16(1):e0246095. doi: 10.1371/journal.pone.0246095. eCollection 2021. PLoS One. 2021. PMID: 33508026 Free PMC article.
-
The Expression of TaRca2-α Gene Associated with Net Photosynthesis Rate, Biomass and Grain Yield in Bread Wheat (Triticum aestivum L.) under Field Conditions.PLoS One. 2016 Aug 22;11(8):e0161308. doi: 10.1371/journal.pone.0161308. eCollection 2016. PLoS One. 2016. PMID: 27548477 Free PMC article.
-
Characterization of the multigene family TaHKT 2;1 in bread wheat and the role of gene members in plant Na(+) and K(+) status.BMC Plant Biol. 2014 Jun 11;14:159. doi: 10.1186/1471-2229-14-159. BMC Plant Biol. 2014. PMID: 24920193 Free PMC article.
References
LinkOut - more resources
Full Text Sources

