Transcript profiling of two alfalfa genotypes with contrasting cell wall composition in stems using a cross-species platform: optimizing analysis by masking biased probes
- PMID: 20497574
- PMCID: PMC2893600
- DOI: 10.1186/1471-2164-11-323
Transcript profiling of two alfalfa genotypes with contrasting cell wall composition in stems using a cross-species platform: optimizing analysis by masking biased probes
Abstract
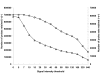
Background: The GeneChip(R) Medicago Genome Array, developed for Medicago truncatula, is a suitable platform for transcript profiling in tetraploid alfalfa [Medicago sativa (L.) subsp. sativa]. However, previous research involving cross-species hybridization (CSH) has shown that sequence variation between two species can bias transcript profiling by decreasing sensitivity (number of expressed genes detected) and the accuracy of measuring fold-differences in gene expression.
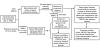
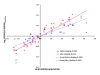
Results: Transcript profiling using the Medicago GeneChip(R) was conducted with elongating stem (ES) and post-elongation stem (PES) internodes from alfalfa genotypes 252 and 1283 that differ in stem cell wall concentrations of cellulose and lignin. A protocol was developed that masked probes targeting inter-species variable (ISV) regions of alfalfa transcripts. A probe signal intensity threshold was selected that optimized both sensitivity and accuracy. After masking for both ISV regions and previously identified single-feature polymorphisms (SFPs), the number of differentially expressed genes between the two genotypes in both ES and PES internodes was approximately 2-fold greater than the number detected prior to masking. Regulatory genes, including transcription factor and receptor kinase genes that may play a role in development of secondary xylem, were significantly over-represented among genes up-regulated in 252 PES internodes compared to 1283 PES internodes. Several cell wall-related genes were also up-regulated in genotype 252 PES internodes. Real-time quantitative RT-PCR of differentially expressed regulatory and cell wall-related genes demonstrated increased sensitivity and accuracy after masking for both ISV regions and SFPs. Over 1,000 genes that were differentially expressed in ES and PES internodes of genotypes 252 and 1283 were mapped onto putative orthologous loci on M. truncatula chromosomes. Clustering simulation analysis of the differentially expressed genes suggested co-expression of some neighbouring genes on Medicago chromosomes.
Conclusions: The problems associated with transcript profiling in alfalfa stems using the Medicago GeneChip as a CSH platform were mitigated by masking probes targeting ISV regions and SFPs. Using this masking protocol resulted in the identification of numerous candidate genes that may contribute to differences in cell wall concentration and composition of stems of two alfalfa genotypes.
Figures






References
-
- Michaud R, Lehman WF, Rumbaugh MD. Alfalfa and alfalfa improvement -- Agronomy Monograph no. 29. Madison, WI: ASA-CSSA-SSSA; 1988. World distribution and historical development; pp. 25–91.
-
- Samac DA, Jung H-JG, Lamb JFS. In: Alcoholic Fuels. Minteer SD, editor. Boca Raton, FL: CRC Press; 2006. Development of alfalfa (Medicago sativa L.) as a feedstock for production of ethanol and other bioproducts; pp. 79–98.
-
- National Agricultural Statistics Service. On-line resource. 2008. http://www.nass.usda.gov
-
- Russelle MP, Birr AS. Large-scale assessment of symbiotic dinitrogen fixation by crops: Soybean and alfalfa in the Mississippi river basin. Agron J. 2004;96:1754–1760.
-
- Angers DA. Changes in soil aggregation and organic carbon under corn and alfalfa. Soil Sci Soc Am J. 1992;56:1244–1249.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

