An unusual function of RON receptor tyrosine kinase as a transcriptional regulator in cooperation with EGFR in human cancer cells
- PMID: 20498137
- PMCID: PMC2915629
- DOI: 10.1093/carcin/bgq100
An unusual function of RON receptor tyrosine kinase as a transcriptional regulator in cooperation with EGFR in human cancer cells
Abstract
Homodimerization of RON (MST1R), a receptor tyrosine kinase, usually occurs in cells stimulated by a ligand and leads to the downstream activation of signaling pathways. Here we report that bladder cancer cells, in response to physiological stress, use an alternative mechanism for signaling activation. Time-course studies indicated that RON migrated directly from the membrane to the nucleus of bladder cancer cells in response to serum starvation. Biochemical and genetic studies implied that this nuclear internalization was complexed with epidermal growth factor receptor (EGFR) and required the docking of importins. In vivo analysis confirmed that nuclear RON was present in 38.4% (28/73) of primary bladder tumors. Chromatin immunoprecipitation (ChIP) on microarray analysis further revealed that this internalized complex bound to at least 134 target genes known to participate in three stress-responsive networks: p53, stress-activated protein kinase/c-jun N-terminal kinase and phosphatidylinositol 3-kinase/Akt. These findings suggest that RON, in a complex with EGFR, acts as a transcriptional regulator in response to acute disturbances (e.g. serum starvation) imposed on cancer cells. In an attempt to re-establish homeostasis, these cells bypass regular mechanisms required by ligand stimulation and trigger the RON-directed transcriptional response, which confers a survival advantage.
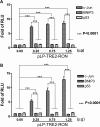
Figures





Similar articles
-
Nuclear translocation of RON receptor tyrosine kinase. New mechanistic and functional insights.Cytokine Growth Factor Rev. 2025 Feb;81:9-15. doi: 10.1016/j.cytogfr.2024.12.004. Epub 2025 Jan 2. Cytokine Growth Factor Rev. 2025. PMID: 39794156 Review.
-
EGFR transactivates RON to drive oncogenic crosstalk.Elife. 2021 Nov 25;10:e63678. doi: 10.7554/eLife.63678. Elife. 2021. PMID: 34821550 Free PMC article.
-
Hypoxia promotes nuclear translocation and transcriptional function in the oncogenic tyrosine kinase RON.Cancer Res. 2014 Aug 15;74(16):4549-62. doi: 10.1158/0008-5472.CAN-13-3730. Epub 2014 Jun 5. Cancer Res. 2014. PMID: 24903148
-
Plasma membrane proteoglycans syndecan-2 and syndecan-4 engage with EGFR and RON kinase to sustain carcinoma cell cycle progression.J Biol Chem. 2022 Jun;298(6):102029. doi: 10.1016/j.jbc.2022.102029. Epub 2022 May 13. J Biol Chem. 2022. PMID: 35569509 Free PMC article.
-
Ron-receptor tyrosine kinase in tumorigenesis and metastasis.Future Oncol. 2007 Aug;3(4):441-8. doi: 10.2217/14796694.3.4.441. Future Oncol. 2007. PMID: 17661719 Free PMC article. Review.
Cited by
-
Dietary restriction: could it be considered as speed bump on tumor progression road?Tumour Biol. 2016 Jun;37(6):7109-18. doi: 10.1007/s13277-016-5044-8. Epub 2016 Apr 4. Tumour Biol. 2016. PMID: 27043958 Review.
-
Evidence for 2-Methoxyestradiol-Mediated Inhibition of Receptor Tyrosine Kinase RON in the Management of Prostate Cancer.Int J Mol Sci. 2021 Feb 12;22(4):1852. doi: 10.3390/ijms22041852. Int J Mol Sci. 2021. PMID: 33673346 Free PMC article.
-
An Introduction and Overview of RON Receptor Tyrosine Kinase Signaling.Genes (Basel). 2023 Feb 17;14(2):517. doi: 10.3390/genes14020517. Genes (Basel). 2023. PMID: 36833444 Free PMC article. Review.
-
Receptor tyrosine kinase recepteur d'origine nantais as predictive marker for aggressive prostate cancer in African Americans.Mol Carcinog. 2019 Jun;58(6):854-861. doi: 10.1002/mc.23002. Epub 2019 Mar 11. Mol Carcinog. 2019. PMID: 30859654 Free PMC article.
-
Ron tyrosine kinase receptor synergises with EGFR to confer adverse features in head and neck squamous cell carcinoma.Br J Cancer. 2013 Jul 23;109(2):482-92. doi: 10.1038/bjc.2013.321. Epub 2013 Jun 25. Br J Cancer. 2013. PMID: 23799848 Free PMC article.
References
-
- Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2000;103:211–225. - PubMed
-
- Sorkin A, et al. Signal transduction and endocytosis: close encounters of many kinds. Nat. Rev. 2002;3:600–614. - PubMed
-
- Gschwind A, et al. Cell communication networks: epidermal growth factor receptor transactivation as the paradigm for inter-receptor signal transmission. Oncogene. 2001;20:1594–1600. - PubMed
-
- Ronsin C, et al. A novel putative receptor protein tyrosine kinase of the met family. Oncogene. 1993;8:1195–1202. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

