BUD22 affects Ty1 retrotransposition and ribosome biogenesis in Saccharomyces cerevisiae
- PMID: 20498295
- PMCID: PMC2927749
- DOI: 10.1534/genetics.110.119115
BUD22 affects Ty1 retrotransposition and ribosome biogenesis in Saccharomyces cerevisiae
Abstract
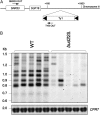
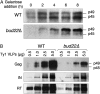
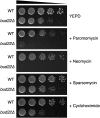
A variety of cellular factors affect the movement of the retrovirus-like transposon Ty1. To identify genes involved in Ty1 virus-like particle (VLP) function, the level of the major capsid protein (Gag-p45) and its proteolytic precursor (Gag-p49p) was monitored in a subset of Ty1 cofactor mutants. Twenty-nine of 87 mutants contained alterations in the level of Gag; however, only bud22Delta showed a striking defect in Gag processing. BUD22 affected the +1 translational frameshifting event required to express the Pol proteins protease, integrase, and reverse transcriptase. Therefore, it is possible that the bud22Delta mutant may not produce enough functional Ty1 protease to completely process Gag-p49 to p45. Furthermore, BUD22 is required for 18S rRNA processing and 40S subunit biogenesis and influences polysome density. Together our results suggest that BUD22 is involved in a step in ribosome biogenesis that not only affects general translation, but also may alter the frameshifting efficiency of ribosomes, an event central to Ty1 retrotransposition.
Figures




Similar articles
-
Ribosome Biogenesis Modulates Ty1 Copy Number Control in Saccharomyces cerevisiae.Genetics. 2017 Dec;207(4):1441-1456. doi: 10.1534/genetics.117.300388. Epub 2017 Oct 18. Genetics. 2017. PMID: 29046400 Free PMC article.
-
Yeast Krr1p physically and functionally interacts with a novel essential Kri1p, and both proteins are required for 40S ribosome biogenesis in the nucleolus.Mol Cell Biol. 2000 Nov;20(21):7971-9. doi: 10.1128/MCB.20.21.7971-7979.2000. Mol Cell Biol. 2000. PMID: 11027267 Free PMC article.
-
The pokeweed antiviral protein specifically inhibits Ty1-directed +1 ribosomal frameshifting and retrotransposition in Saccharomyces cerevisiae.J Virol. 1998 Feb;72(2):1036-42. doi: 10.1128/JVI.72.2.1036-1042.1998. J Virol. 1998. PMID: 9444997 Free PMC article.
-
Ribosome biogenesis factors-from names to functions.EMBO J. 2023 Apr 3;42(7):e112699. doi: 10.15252/embj.2022112699. Epub 2023 Feb 10. EMBO J. 2023. PMID: 36762427 Free PMC article. Review.
-
Mitochondrial ribosomal protein genes connected with Alzheimer's and tellurite toxicity.Mitochondrion. 2022 May;64:45-58. doi: 10.1016/j.mito.2022.02.006. Epub 2022 Feb 24. Mitochondrion. 2022. PMID: 35218961 Review.
Cited by
-
An interchangeable prion-like domain is required for Ty1 retrotransposition.bioRxiv [Preprint]. 2023 Feb 27:2023.02.27.530227. doi: 10.1101/2023.02.27.530227. bioRxiv. 2023. Update in: Proc Natl Acad Sci U S A. 2023 Jul 25;120(30):e2303358120. doi: 10.1073/pnas.2303358120. PMID: 36909481 Free PMC article. Updated. Preprint.
-
Ribosomal protein and biogenesis factors affect multiple steps during movement of the Saccharomyces cerevisiae Ty1 retrotransposon.Mob DNA. 2015 Dec 8;6:22. doi: 10.1186/s13100-015-0053-5. eCollection 2015. Mob DNA. 2015. PMID: 26664557 Free PMC article.
-
Lifespan Extension by Retrotransposons under Conditions of Mild Stress Requires Genes Involved in tRNA Modifications and Nucleotide Metabolism.Int J Mol Sci. 2024 Oct 1;25(19):10593. doi: 10.3390/ijms251910593. Int J Mol Sci. 2024. PMID: 39408922 Free PMC article.
-
A nuclear pore sub-complex restricts the propagation of Ty retrotransposons by limiting their transcription.PLoS Genet. 2021 Nov 1;17(11):e1009889. doi: 10.1371/journal.pgen.1009889. eCollection 2021 Nov. PLoS Genet. 2021. PMID: 34723966 Free PMC article.
-
Control of yeast retrotransposons mediated through nucleoporin evolution.PLoS Genet. 2018 Apr 25;14(4):e1007325. doi: 10.1371/journal.pgen.1007325. eCollection 2018 Apr. PLoS Genet. 2018. PMID: 29694349 Free PMC article.
References
-
- Adams, S. E., J. Mellor, K. Gull, R. B. Sim, M. F. Tuite et al., 1987. The functions and relationships of Ty-VLP proteins in yeast reflect those of mammalian retroviral proteins. Cell 49 111–119. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

