Serpins flex their muscle: II. Structural insights into target peptidase recognition, polymerization, and transport functions
- PMID: 20498368
- PMCID: PMC2915666
- DOI: 10.1074/jbc.R110.141408
Serpins flex their muscle: II. Structural insights into target peptidase recognition, polymerization, and transport functions
Abstract
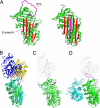
Inhibitory serpins are metastable proteins that undergo a substantial conformational rearrangement to covalently trap target peptidases. The serpin reactive center loop contributes a majority of the interactions that serpins make during the initial binding to target peptidases. However, structural studies on serpin-peptidase complexes reveal a broader set of contacts on the scaffold of inhibitory serpins that have substantial influence on guiding peptidase recognition. Structural and biophysical studies also reveal how aberrant serpin folding can lead to the formation of domain-swapped serpin multimers rather than the monomeric metastable state. Serpin domain swapping may therefore underlie the polymerization events characteristic of the serpinopathies. Finally, recent structural studies reveal how the serpin fold has been adapted for non-inhibitory functions such as hormone binding.
Figures

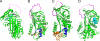
Similar articles
-
Reactive centre loop dynamics and serpin specificity.Sci Rep. 2019 Mar 7;9(1):3870. doi: 10.1038/s41598-019-40432-w. Sci Rep. 2019. PMID: 30846766 Free PMC article.
-
A structure-derived snap-trap mechanism of a multispecific serpin from the dysbiotic human oral microbiome.J Biol Chem. 2017 Jun 30;292(26):10883-10898. doi: 10.1074/jbc.M117.786533. Epub 2017 May 16. J Biol Chem. 2017. PMID: 28512127 Free PMC article.
-
Inhibitory serpins. New insights into their folding, polymerization, regulation and clearance.Biochem J. 2016 Aug 1;473(15):2273-93. doi: 10.1042/BCJ20160014. Biochem J. 2016. PMID: 27470592 Free PMC article. Review.
-
Analysis of surface cavity in serpin family reveals potential binding sites for chemical chaperone to reduce polymerization.J Mol Model. 2012 Mar;18(3):1143-51. doi: 10.1007/s00894-011-1110-8. Epub 2011 Jun 17. J Mol Model. 2012. PMID: 21681443
-
A protein family under 'stress' - serpin stability, folding and misfolding.Front Biosci. 2005 Jan 1;10:288-99. doi: 10.2741/1528. Print 2005 Jan 1. Front Biosci. 2005. PMID: 15574369 Review.
Cited by
-
The circadian Clock gene regulates acrosin activity of sperm through serine protease inhibitor A3K.Exp Biol Med (Maywood). 2016 Jan;241(2):205-15. doi: 10.1177/1535370215597199. Epub 2015 Aug 11. Exp Biol Med (Maywood). 2016. PMID: 26264441 Free PMC article.
-
Conformational transition of the Ixodes ricinus salivary serpin Iripin-4.Acta Crystallogr D Struct Biol. 2023 May 1;79(Pt 5):409-419. doi: 10.1107/S2059798323002322. Epub 2023 Apr 24. Acta Crystallogr D Struct Biol. 2023. PMID: 37092969 Free PMC article.
-
SERPINB12 Is a Slow-Binding Inhibitor of Granzyme A and Hepsin.Biochemistry. 2015 Nov 17;54(45):6756-9. doi: 10.1021/acs.biochem.5b01042. Epub 2015 Nov 5. Biochemistry. 2015. PMID: 26497600 Free PMC article.
-
Ixodes ricinus Salivary Serpin Iripin-8 Inhibits the Intrinsic Pathway of Coagulation and Complement.Int J Mol Sci. 2021 Aug 31;22(17):9480. doi: 10.3390/ijms22179480. Int J Mol Sci. 2021. PMID: 34502392 Free PMC article.
-
The effects of PEDF on cancer biology: mechanisms of action and therapeutic potential.Nat Rev Cancer. 2013 Apr;13(4):258-71. doi: 10.1038/nrc3484. Epub 2013 Mar 14. Nat Rev Cancer. 2013. PMID: 23486238 Free PMC article. Review.
References
-
- Irving J. A., Pike R. N., Lesk A. M., Whisstock J. C. (2000) Genome Res. 10, 1845–1864 - PubMed
-
- Huntington J. A. (2006) Trends Biochem. Sci. 31, 427–435 - PubMed
-
- Huntington J. A., Read R. J., Carrell R. W. (2000) Nature 407, 923–926 - PubMed
-
- Whisstock J. C., Bottomley S. P. (2006) Curr. Opin. Struct. Biol. 16, 761–768 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

