On the significance of an RNA tertiary structure prediction
- PMID: 20498460
- PMCID: PMC2885683
- DOI: 10.1261/rna.1837410
On the significance of an RNA tertiary structure prediction
Abstract
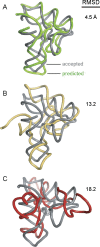
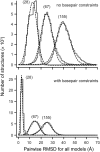
Tertiary structure prediction is important for understanding structure-function relationships for RNAs whose structures are unknown and for characterizing RNA states recalcitrant to direct analysis. However, it is unknown what root-mean-square deviation (RMSD) corresponds to a statistically significant RNA tertiary structure prediction. We use discrete molecular dynamics to generate RNA-like folds for structures up to 161 nucleotides (nt) that have complex tertiary interactions and then determine the RMSD distribution between these decoys. These distributions are Gaussian-like. The mean RMSD increases with RNA length and is smaller if secondary structure constraints are imposed while generating decoys. The compactness of RNA molecules with true tertiary folds is intermediate between closely packed spheres and a freely jointed chain. We use this scaling relationship to define an expression relating RMSD with the confidence that a structure prediction is better than that expected by chance. This is the prediction significance, and corresponds to a P-value. For a 100-nt RNA, the RMSD of predicted structures should be within 25 A of the accepted structure to reach the P <or= 0.01 level if the secondary structure is predicted de novo and within 14 A if secondary structure information is used as a constraint. This significance approach should be useful for evaluating diverse RNA structure prediction and molecular modeling algorithms.
Figures




References
-
- Batey RT, Gilbert SD, Montange RK 2004. Structure of a natural guanine-responsive riboswitch complexed with the metabolite hypoxanthine. Nature 432: 411–415 - PubMed
-
- Cohen F, Sternberg MJE 1980. On the prediction of protein structure: The significance of the root-mean-square deviation. J Mol Biol 138: 321–333 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
