Using population genetic theory and DNA sequences for species detection and identification in asexual organisms
- PMID: 20498705
- PMCID: PMC2869354
- DOI: 10.1371/journal.pone.0010609
Using population genetic theory and DNA sequences for species detection and identification in asexual organisms
Abstract
Background: It is widely agreed that species are fundamental units of biology, but there is little agreement on a definition of species or on an operational criterion for delimiting species that is applicable to all organisms.
Methodology/principal findings: We focus on asexual eukaryotes as the simplest case for investigating species and speciation. We describe a model of speciation in asexual organisms based on basic principles of population and evolutionary genetics. The resulting species are independently evolving populations as described by the evolutionary species concept or the general lineage species concept. Based on this model, we describe a procedure for using gene sequences from small samples of individuals to assign them to the same or different species. Using this method of species delimitation, we demonstrate the existence of species as independent evolutionary units in seven groups of invertebrates, fungi, and protists that reproduce asexually most or all of the time.
Conclusions/significance: This wide evolutionary sampling establishes the general existence of species and speciation in asexual organisms. The method is well suited for measuring species diversity when phenotypic data are insufficient to distinguish species, or are not available, as in DNA barcoding and environmental sequencing. We argue that it is also widely applicable to sexual organisms.
Conflict of interest statement
Figures


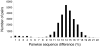
References
-
- Darwin C. London: 1872. The Origin of Species by Means of Natural Selection. John Murray.
-
- Howard DJ, Berlocher SH, editors. New York, New York: Oxford University Press; 1999. Endless Forms.
-
- Wheeler Q, Meier R. New York: Columbia University Press; 2000. Species Concepts and Phylogenetic Theory.
-
- Coyne JA, Orr HA. Sunderland, Massachusetts: Sinauer Associates, Inc; 2004. Speciation.
-
- de Queiroz K. Species concepts and species delimitation. Syst Biol. 2007;56:879–886. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources

