Searching for signaling balance through the identification of genetic interactors of the Rab guanine-nucleotide dissociation inhibitor gdi-1
- PMID: 20498707
- PMCID: PMC2869356
- DOI: 10.1371/journal.pone.0010624
Searching for signaling balance through the identification of genetic interactors of the Rab guanine-nucleotide dissociation inhibitor gdi-1
Abstract
Background: The symptoms of numerous diseases result from genetic mutations that disrupt the homeostasis maintained by the appropriate integration of signaling gene activities. The relationships between signaling genes suggest avenues through which homeostasis can be restored and disease symptoms subsequently reduced. Specifically, disease symptoms caused by loss-of-function mutations in a particular gene may be reduced by concomitant perturbations in genes with antagonistic activities.
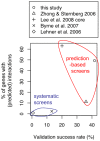
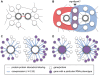
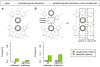
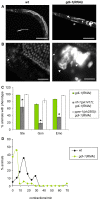
Methodology/principal findings: Here we use network-neighborhood analyses to predict genetic interactions in Caenorhabditis elegans towards mapping antagonisms and synergisms between genes in an animal model. Most of the predicted interactions are novel, and the experimental validation establishes that our approach provides a gain in accuracy compared to previous efforts. In particular, we identified genetic interactors of gdi-1, the orthologue of GDI1, a gene associated with mental retardation in human. Interestingly, some gdi-1 interactors have human orthologues with known neurological functions, and upon validation of the interactions in mammalian systems, these orthologues would be potential therapeutic targets for GDI1-associated neurological disorders. We also observed the conservation of a gdi-1 interaction between different cellular systems in C. elegans, suggesting the involvement of GDI1 in human muscle degeneration.
Conclusions/significance: We developed a novel predictor of genetic interactions that may have the ability to significantly streamline the identification of therapeutic targets for monogenic disorders involving genes conserved between human and C. elegans.
Conflict of interest statement
Figures

Similar articles
-
A Sensitized Screen for Genes Promoting Invadopodia Function In Vivo: CDC-42 and Rab GDI-1 Direct Distinct Aspects of Invadopodia Formation.PLoS Genet. 2016 Jan 14;12(1):e1005786. doi: 10.1371/journal.pgen.1005786. eCollection 2016 Jan. PLoS Genet. 2016. PMID: 26765257 Free PMC article.
-
Cortical localization of the Galpha protein GPA-16 requires RIC-8 function during C. elegans asymmetric cell division.Development. 2005 Oct;132(20):4449-59. doi: 10.1242/dev.02039. Epub 2005 Sep 14. Development. 2005. PMID: 16162648
-
Regulation of Caenorhabditis elegans RNA interference by the daf-2 insulin stress and longevity signaling pathway.Cold Spring Harb Symp Quant Biol. 2004;69:429-31. doi: 10.1101/sqb.2004.69.429. Cold Spring Harb Symp Quant Biol. 2004. PMID: 16117677 No abstract available.
-
Networks in Caenorhabditis elegans.Curr Opin Genet Dev. 2011 Dec;21(6):787-98. doi: 10.1016/j.gde.2011.10.003. Epub 2011 Nov 4. Curr Opin Genet Dev. 2011. PMID: 22054717 Review.
-
A Caenorhabditis elegans genetic-interaction map wiggles into view.J Biol. 2008;7(3):8. doi: 10.1186/jbiol70. Epub 2008 Mar 7. J Biol. 2008. PMID: 18341704 Free PMC article. Review.
Cited by
-
Restoring de novo coenzyme Q biosynthesis in Caenorhabditis elegans coq-3 mutants yields profound rescue compared to exogenous coenzyme Q supplementation.Gene. 2012 Sep 10;506(1):106-16. doi: 10.1016/j.gene.2012.06.023. Epub 2012 Jun 23. Gene. 2012. PMID: 22735617 Free PMC article.
-
Zyxin contributes to coupling between cell junctions and contractile actomyosin networks during apical constriction.PLoS Genet. 2023 Mar 28;19(3):e1010319. doi: 10.1371/journal.pgen.1010319. eCollection 2023 Mar. PLoS Genet. 2023. PMID: 36976799 Free PMC article.
-
Structural and Functional Characterization of a Caenorhabditis elegans Genetic Interaction Network within Pathways.PLoS Comput Biol. 2016 Feb 12;12(2):e1004738. doi: 10.1371/journal.pcbi.1004738. eCollection 2016 Feb. PLoS Comput Biol. 2016. PMID: 26871911 Free PMC article.
-
Beyond Synthetic Lethality: Charting the Landscape of Pairwise Gene Expression States Associated with Survival in Cancer.Cell Rep. 2019 Jul 23;28(4):938-948.e6. doi: 10.1016/j.celrep.2019.06.067. Cell Rep. 2019. PMID: 31340155 Free PMC article.
-
Caenorhabditis elegans as a Model System for Duchenne Muscular Dystrophy.Int J Mol Sci. 2021 May 5;22(9):4891. doi: 10.3390/ijms22094891. Int J Mol Sci. 2021. PMID: 34063069 Free PMC article. Review.
References
-
- Schubert V, Dotti CG. Transmitting on actin: synaptic control of dendritic architecture. J Cell Sci. 2007;120:205–212. - PubMed
-
- Newey SE, Velamoor V, Govek EE, Van Aelst L. Rho GTPases, dendritic structure, and mental retardation. J Neurobiol. 2005;64:58–74. - PubMed
-
- Kutsche K, Yntema H, Brandt A, Jantke I, Nothwang HG, et al. Mutations in ARHGEF6, encoding a guanine nucleotide exchange factor for Rho GTPases, in patients with X-linked mental retardation. Nat Genet. 2000;26:247–250. - PubMed
-
- Tavazoie SF, Alvarez VA, Ridenour DA, Kwiatkowski DJ, Sabatini BL. Regulation of neuronal morphology and function by the tumor suppressors Tsc1 and Tsc2. Nat Neurosci. 2005;8:1727–1734. - PubMed
-
- D'Adamo P, Menegon A, Lo Nigro C, Grasso M, Gulisano M, et al. Mutations in GDI1 are responsible for X-linked non-specific mental retardation. Nat Genet. 1998;19:134–139. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

