SETDB1 is involved in postembryonic DNA methylation and gene silencing in Drosophila
- PMID: 20498723
- PMCID: PMC2871795
- DOI: 10.1371/journal.pone.0010581
SETDB1 is involved in postembryonic DNA methylation and gene silencing in Drosophila
Retraction in
-
Retraction: SETDB1 Is Involved in Postembryonic DNA Methylation and Gene Silencing in Drosophila.PLoS One. 2018 Mar 20;13(3):e0194869. doi: 10.1371/journal.pone.0194869. eCollection 2018. PLoS One. 2018. PMID: 29558503 Free PMC article. No abstract available.
Abstract
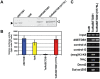
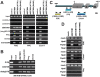
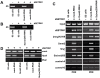
DNA methylation is fundamental for the stability and activity of genomes. Drosophila melanogaster and vertebrates establish a global DNA methylation pattern of their genome during early embryogenesis. Large-scale analyses of DNA methylation patterns have uncovered revealed that DNA methylation patterns are dynamic rather than static and change in a gene-specific fashion during development and in diseased cells. However, the factors and mechanisms involved in dynamic, postembryonic DNA methylation remain unclear. Methylation of lysine 9 in histone H3 (H3-K9) by members of the Su(var)3-9 family of histone methyltransferases (HMTs) triggers embryonic DNA methylation in Arthropods and Chordates. Here, we demonstrate that Drosophila SETDB1 (dSETDB1) can mediate DNA methylation and silencing of genes and retrotransposons. We found that dSETDB1 tri-methylates H3-K9 and binds methylated CpA motifs. Tri-methylation of H3-K9 by dSETDB1 mediates recruitment of DNA methyltransferase 2 (Dnmt2) and Su(var)205, the Drosophila ortholog of mammalian "Heterochromatin Protein 1", to target genes for dSETDB1. By enlisting Dnmt2 and Su(var)205, dSETDB1 triggers DNA methylation and silencing of genes and retrotransposons in Drosophila cells. DSETDB1 is involved in postembryonic DNA methylation and silencing of Rt1b{} retrotransposons and the tumor suppressor gene retinoblastoma family protein 1 (Rb) in imaginal discs. Collectively, our findings implicate dSETDB1 in postembryonic DNA methylation, provide a model for silencing of the tumor suppressor Rb, and uncover a role for cell type-specific DNA methylation in Drosophila development.
Conflict of interest statement
Figures


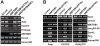




Comment in
-
Findings of Research Misconduct.Fed Regist. 2017 Jul 6;82(128):31334-31335. Fed Regist. 2017. PMID: 28752862 Free PMC article. No abstract available.
References
-
- Bird AP, Wolffe AP. Methylation-induced repression – belts, braces, and chromatin. Cell. 1999;99:451–454. - PubMed
-
- Wilson GG, Murray NE. Restriction and modification systems. Annu Rev Genet. 1991;25:585–627. - PubMed
-
- Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;Suppl 33:245–254. - PubMed
-
- Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet. 2008;9:465–476. - PubMed
-
- Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem Sci. 2006;31:89–97. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

