Re-annotation is an essential step in systems biology modeling of functional genomics data
- PMID: 20498845
- PMCID: PMC2871057
- DOI: 10.1371/journal.pone.0010642
Re-annotation is an essential step in systems biology modeling of functional genomics data
Abstract
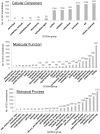
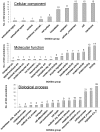
One motivation of systems biology research is to understand gene functions and interactions from functional genomics data such as that derived from microarrays. Up-to-date structural and functional annotations of genes are an essential foundation of systems biology modeling. We propose that the first essential step in any systems biology modeling of functional genomics data, especially for species with recently sequenced genomes, is gene structural and functional re-annotation. To demonstrate the impact of such re-annotation, we structurally and functionally re-annotated a microarray developed, and previously used, as a tool for disease research. We quantified the impact of this re-annotation on the array based on the total numbers of structural- and functional-annotations, the Gene Annotation Quality (GAQ) score, and canonical pathway coverage. We next quantified the impact of re-annotation on systems biology modeling using a previously published experiment that used this microarray. We show that re-annotation improves the quantity and quality of structural- and functional-annotations, allows a more comprehensive Gene Ontology based modeling, and improves pathway coverage for both the whole array and a differentially expressed mRNA subset. Our results also demonstrate that re-annotation can result in a different knowledge outcome derived from previous published research findings. We propose that, because of this, re-annotation should be considered to be an essential first step for deriving value from functional genomics data.
Conflict of interest statement
Figures
References
-
- Schena M, Heller RA, Theriault TP, Konrad K, Lachenmeier E, et al. Microarrays: biotechnology's discovery platform for functional genomics. Trends Biotechnol. 1998;16:301–306. - PubMed
-
- Sellheyer K, Belbin TJ. DNA microarrays: from structural genomics to functional genomics. The applications of gene chips in dermatology and dermatopathology. J Am Acad Dermatol. 2004;51:681–692; quiz 693-686. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

