Expansion of ribosomally produced natural products: a nitrile hydratase- and Nif11-related precursor family
- PMID: 20500830
- PMCID: PMC2887384
- DOI: 10.1186/1741-7007-8-70
Expansion of ribosomally produced natural products: a nitrile hydratase- and Nif11-related precursor family
Abstract
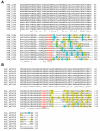
Background: A new family of natural products has been described in which cysteine, serine and threonine from ribosomally-produced peptides are converted to thiazoles, oxazoles and methyloxazoles, respectively. These metabolites and their biosynthetic gene clusters are now referred to as thiazole/oxazole-modified microcins (TOMM). As exemplified by microcin B17 and streptolysin S, TOMM precursors contain an N-terminal leader sequence and C-terminal core peptide. The leader sequence contains binding sites for the posttranslational modifying enzymes which subsequently act upon the core peptide. TOMM peptides are small and highly variable, frequently missed by gene-finders and occasionally situated far from the thiazole/oxazole forming genes. Thus, locating a substrate for a particular TOMM pathway can be a challenging endeavor.
Results: Examination of candidate TOMM precursors has revealed a subclass with an uncharacteristically long leader sequence closely related to the enzyme nitrile hydratase. Members of this nitrile hydratase leader peptide (NHLP) family lack the metal-binding residues required for catalysis. Instead, NHLP sequences display the classic Gly-Gly cleavage motif and have C-terminal regions rich in heterocyclizable residues. The NHLP family exhibits a correlated species distribution and local clustering with an ABC transport system. This study also provides evidence that a separate family, annotated as Nif11 nitrogen-fixing proteins, can serve as natural product precursors (N11P), but not always of the TOMM variety. Indeed, a number of cyanobacterial genomes show extensive N11P paralogous expansion, such as Nostoc, Prochlorococcus and Cyanothece, which replace the TOMM cluster with lanthionine biosynthetic machinery.
Conclusions: This study has united numerous TOMM gene clusters with their cognate substrates. These results suggest that two large protein families, the nitrile hydratases and Nif11, have been retailored for secondary metabolism. Precursors for TOMMs and lanthionine-containing peptides derived from larger proteins to which other functions are attributed, may be widespread. The functions of these natural products have yet to be elucidated, but it is probable that some will display valuable industrial or medical activities.
Figures




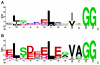
Comment in
-
The hidden diversity of ribosomal peptide natural products.BMC Biol. 2010 Jun 17;8:83. doi: 10.1186/1741-7007-8-83. BMC Biol. 2010. PMID: 20594290 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

