Metabolic flexibility revealed in the genome of the cyst-forming alpha-1 proteobacterium Rhodospirillum centenum
- PMID: 20500872
- PMCID: PMC2890560
- DOI: 10.1186/1471-2164-11-325
Metabolic flexibility revealed in the genome of the cyst-forming alpha-1 proteobacterium Rhodospirillum centenum
Abstract
Background: Rhodospirillum centenum is a photosynthetic non-sulfur purple bacterium that favors growth in an anoxygenic, photosynthetic N2-fixing environment. It is emerging as a genetically amenable model organism for molecular genetic analysis of cyst formation, photosynthesis, phototaxis, and cellular development. Here, we present an analysis of the genome of this bacterium.
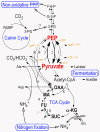
Results: R. centenum contains a singular circular chromosome of 4,355,548 base pairs in size harboring 4,105 genes. It has an intact Calvin cycle with two forms of Rubisco, as well as a gene encoding phosphoenolpyruvate carboxylase (PEPC) for mixotrophic CO2 fixation. This dual carbon-fixation system may be required for regulating internal carbon flux to facilitate bacterial nitrogen assimilation. Enzymatic reactions associated with arsenate and mercuric detoxification are rare or unique compared to other purple bacteria. Among numerous newly identified signal transduction proteins, of particular interest is a putative bacteriophytochrome that is phylogenetically distinct from a previously characterized R. centenum phytochrome, Ppr. Genes encoding proteins involved in chemotaxis as well as a sophisticated dual flagellar system have also been mapped.
Conclusions: Remarkable metabolic versatility and a superior capability for photoautotrophic carbon assimilation is evident in R. centenum.
Figures




References
-
- Stadtwald-Demchick R, Turner FR, Gest H. Physiological properties of the thermotolerant photosynthetic bacterium, Rhodospirillum centenum. FEMS Microbiol Lett. 1990;11:139–144. doi: 10.1111/j.1574-6968.1990.tb13851.x. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

