Genetic structure of the Spanish population
- PMID: 20500880
- PMCID: PMC2887418
- DOI: 10.1186/1471-2164-11-326
Genetic structure of the Spanish population
Abstract
Background: Genetic admixture is a common caveat for genetic association analysis. Therefore, it is important to characterize the genetic structure of the population under study to control for this kind of potential bias.
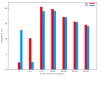
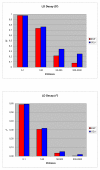
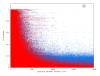
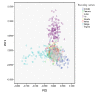
Results: In this study we have sampled over 800 unrelated individuals from the population of Spain, and have genotyped them with a genome-wide coverage. We have carried out linkage disequilibrium, haplotype, population structure and copy-number variation (CNV) analyses, and have compared these estimates of the Spanish population with existing data from similar efforts.
Conclusions: In general, the Spanish population is similar to the Western and Northern Europeans, but has a more diverse haplotypic structure. Moreover, the Spanish population is also largely homogeneous within itself, although patterns of micro-structure may be able to predict locations of origin from distant regions. Finally, we also present the first characterization of a CNV map of the Spanish population. These results and original data are made available to the scientific community.
Figures





References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

