CyloFold: secondary structure prediction including pseudoknots
- PMID: 20501603
- PMCID: PMC2896150
- DOI: 10.1093/nar/gkq432
CyloFold: secondary structure prediction including pseudoknots
Abstract
Computational RNA secondary structure prediction approaches differ by the way RNA pseudoknot interactions are handled. For reasons of computational efficiency, most approaches only allow a limited class of pseudoknot interactions or are not considering them at all. Here we present a computational method for RNA secondary structure prediction that is not restricted in terms of pseudoknot complexity. The approach is based on simulating a folding process in a coarse-grained manner by choosing helices based on established energy rules. The steric feasibility of the chosen set of helices is checked during the folding process using a highly coarse-grained 3D model of the RNA structures. Using two data sets of 26 and 241 RNA sequences we find that this approach is competitive compared to the existing RNA secondary structure prediction programs pknotsRG, HotKnots and UnaFold. The key advantages of the new method are that there is no algorithmic restriction in terms of pseudoknot complexity and a test is made for steric feasibility.
Availability: The program is available as web server at the site: http://cylofold.abcc.ncifcrf.gov.
Figures
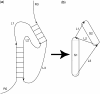

Similar articles
-
pknotsRG: RNA pseudoknot folding including near-optimal structures and sliding windows.Nucleic Acids Res. 2007 Jul;35(Web Server issue):W320-4. doi: 10.1093/nar/gkm258. Epub 2007 May 3. Nucleic Acids Res. 2007. PMID: 17478505 Free PMC article.
-
HotKnots: heuristic prediction of RNA secondary structures including pseudoknots.RNA. 2005 Oct;11(10):1494-504. doi: 10.1261/rna.7284905. RNA. 2005. PMID: 16199760 Free PMC article.
-
DotKnot: pseudoknot prediction using the probability dot plot under a refined energy model.Nucleic Acids Res. 2010 Apr;38(7):e103. doi: 10.1093/nar/gkq021. Epub 2010 Jan 31. Nucleic Acids Res. 2010. PMID: 20123730 Free PMC article.
-
Pseudoknots in RNA Structure Prediction.Curr Protoc. 2023 Feb;3(2):e661. doi: 10.1002/cpz1.661. Curr Protoc. 2023. PMID: 36779804 Review.
-
Beyond Mfold: recent advances in RNA bioinformatics.J Biotechnol. 2006 Jun 25;124(1):41-55. doi: 10.1016/j.jbiotec.2006.01.034. Epub 2006 Mar 10. J Biotechnol. 2006. PMID: 16530285 Free PMC article. Review.
Cited by
-
MPGAfold in dengue secondary structure prediction.Methods Mol Biol. 2014;1138:199-224. doi: 10.1007/978-1-4939-0348-1_13. Methods Mol Biol. 2014. PMID: 24696339 Free PMC article.
-
Sense overlapping transcripts in IS1341-type transposase genes are functional non-coding RNAs in archaea.RNA Biol. 2015;12(5):490-500. doi: 10.1080/15476286.2015.1019998. RNA Biol. 2015. PMID: 25806405 Free PMC article.
-
Bioinformatics of prokaryotic RNAs.RNA Biol. 2014;11(5):470-83. doi: 10.4161/rna.28647. Epub 2014 Apr 2. RNA Biol. 2014. PMID: 24755880 Free PMC article. Review.
-
Possible involvement of three-stemmed pseudoknots in regulating translational initiation in human mRNAs.PLoS One. 2024 Jul 22;19(7):e0307541. doi: 10.1371/journal.pone.0307541. eCollection 2024. PLoS One. 2024. PMID: 39038036 Free PMC article.
-
FMRP regulates actin filament organization via the armadillo protein p0071.RNA. 2013 Nov;19(11):1483-96. doi: 10.1261/rna.037945.112. Epub 2013 Sep 23. RNA. 2013. PMID: 24062571 Free PMC article.
References
-
- Nussinov R, Pieczenik G, Griggs JR, Kleitman DJ. Algorithms for loop matchings. SIAM J. Appl. Math. 1978;35:68–82.
-
- Hofacker IL, Fontana W, Stadler PF, Bonhoeffer S, Tacker M, Schuster P. Fast folding and comparison of RNA secondary structures. Monatshefte f. Chemie. 1994;125:167–188.
-
- Mathews DH, Sabina J, Zuker M, Turner DH. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 1999;288:911–940. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

