Role of peroxisome proliferator-activated receptor-gamma and its coactivator DRIP205 in cellular responses to CDDO (RTA-401) in acute myelogenous leukemia
- PMID: 20501850
- PMCID: PMC3727426
- DOI: 10.1158/0008-5472.CAN-09-1962
Role of peroxisome proliferator-activated receptor-gamma and its coactivator DRIP205 in cellular responses to CDDO (RTA-401) in acute myelogenous leukemia
Abstract
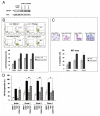
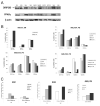
Peroxisome proliferator-activated receptor-gamma (PPARgamma) is a member of the nuclear receptor (NR) family of transcription factors with important regulatory roles in cellular growth, differentiation, and apoptosis. Using proteomic analysis, we showed expression of PPARgamma protein in a series of 260 newly diagnosed primary acute myelogenous leukemia (AML) samples. Forced expression of PPARgamma enhanced the sensitivity of myeloid leukemic cells to apoptosis induced by PPARgamma agonists 2-cyano-3,12-dioxooleana-1,9-dien-28-oic acid (CDDO) and 15-deoxy-(12,14)-15DPGJ(2), through preferential cleavage of caspase-8. No effects on cell cycle distribution or differentiation were noted, despite prominent induction of p21 in PPARgamma-transfected cells. In turn, antagonizing PPARgamma function by small interfering RNA or pharmacologic PPARgamma inhibitor significantly diminished apoptosis induction by CDDO. Overexpression of coactivator protein DRIP205 resulted in enhanced differentiation induction by CDDO in AML cells through PPARgamma activation. Studies with DRIP205 deletion constructs showed that the NR boxes of DRIP205 are not required for this coactivation. In a phase I clinical trial of CDDO (RTA-401) in leukemia, CDDO induced an increase in PPARgamma mRNA expression in six of nine patient samples; of those, induction of differentiation was documented in four patients and that of p21 in three patients, all expressing DRIP205 protein. In summary, these findings suggest that cellular levels of PPARgamma regulate induction of apoptosis via caspase-8 activation, whereas the coactivator DRIP205 is a determinant of induction of differentiation, in response to PPARgamma agonists in leukemic cells.
Figures




Similar articles
-
PPARgamma-active triterpenoid CDDO enhances ATRA-induced differentiation in APL.Cancer Biol Ther. 2007 Dec;6(12):1967-77. doi: 10.4161/cbt.6.12.4982. Epub 2007 Sep 4. Cancer Biol Ther. 2007. PMID: 18075297
-
Peroxisome proliferator-activated receptor gamma and retinoid X receptor ligands are potent inducers of differentiation and apoptosis in leukemias.Mol Cancer Ther. 2004 Oct;3(10):1249-62. Mol Cancer Ther. 2004. PMID: 15486192
-
Activation of peroxisome proliferator-activated receptor gamma by a novel synthetic triterpenoid 2-cyano-3,12-dioxooleana-1,9-dien-28-oic acid induces growth arrest and apoptosis in breast cancer cells.Cancer Res. 2003 Sep 15;63(18):5926-39. Cancer Res. 2003. PMID: 14522919
-
2-Cyano-3,12-dioxoolean-1,9-dien-28-oic acid and related compounds inhibit growth of colon cancer cells through peroxisome proliferator-activated receptor gamma-dependent and -independent pathways.Mol Pharmacol. 2005 Jul;68(1):119-28. doi: 10.1124/mol.105.011437. Epub 2005 Mar 29. Mol Pharmacol. 2005. PMID: 15798084
-
PPARγ Modulators in Lung Cancer: Molecular Mechanisms, Clinical Prospects, and Challenges.Biomolecules. 2024 Feb 4;14(2):190. doi: 10.3390/biom14020190. Biomolecules. 2024. PMID: 38397426 Free PMC article. Review.
Cited by
-
Can an oral antidiabetic (rosiglitazone) be of benefit in leukemia treatment?Saudi Pharm J. 2015 Jan;23(1):14-21. doi: 10.1016/j.jsps.2013.12.009. Epub 2013 Dec 22. Saudi Pharm J. 2015. PMID: 25685038 Free PMC article.
-
Development of a robust classifier for quality control of reverse-phase protein arrays.Bioinformatics. 2015 Mar 15;31(6):912-8. doi: 10.1093/bioinformatics/btu736. Epub 2014 Nov 6. Bioinformatics. 2015. PMID: 25380958 Free PMC article.
-
Safety, pharmacokinetics, and pharmacodynamics of oral omaveloxolone (RTA 408), a synthetic triterpenoid, in a first-in-human trial of patients with advanced solid tumors.Onco Targets Ther. 2017 Aug 29;10:4239-4250. doi: 10.2147/OTT.S136992. eCollection 2017. Onco Targets Ther. 2017. PMID: 28919776 Free PMC article.
-
DIOL triterpenes block profibrotic effects of angiotensin II and protect from cardiac hypertrophy.PLoS One. 2012;7(7):e41545. doi: 10.1371/journal.pone.0041545. Epub 2012 Jul 23. PLoS One. 2012. PMID: 22844495 Free PMC article.
-
Phase I study of the synthetic triterpenoid, 2-cyano-3, 12-dioxoolean-1, 9-dien-28-oic acid (CDDO), in advanced solid tumors.Cancer Chemother Pharmacol. 2012 Feb;69(2):431-8. doi: 10.1007/s00280-011-1712-y. Epub 2011 Jul 31. Cancer Chemother Pharmacol. 2012. PMID: 21805353 Free PMC article. Clinical Trial.
References
-
- Nuclear Receptors Nomenclature Committee A unified nomenclature system for the nuclear receptor superfamily. Cell. 1999;97:161–3. - PubMed
-
- Fajas L, Auboeuf D, Raspe E, et al. The organization, promoter analysis, and expression of the human PPARgamma gene. J Biol Chem. 1997;272:18779–89. - PubMed
-
- Urahama N, Ito M, Sada A, et al. The role of transcriptional coactivator TRAP220 in myelomonocytic differentiation. Genes Cells. 2005;10:1127–37. - PubMed
-
- Hawker NP, Pennypacker SD, Chang SM, Bikle DD. Regulation of human epidermal keratinocyte differentiation by the vitamin D receptor and its coactivators DRIP205, SRC2, and SRC3. J Invest Dermatol. 2007;127:874–80. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

