Most "dark matter" transcripts are associated with known genes
- PMID: 20502517
- PMCID: PMC2872640
- DOI: 10.1371/journal.pbio.1000371
Most "dark matter" transcripts are associated with known genes
Abstract
A series of reports over the last few years have indicated that a much larger portion of the mammalian genome is transcribed than can be accounted for by currently annotated genes, but the quantity and nature of these additional transcripts remains unclear. Here, we have used data from single- and paired-end RNA-Seq and tiling arrays to assess the quantity and composition of transcripts in PolyA+ RNA from human and mouse tissues. Relative to tiling arrays, RNA-Seq identifies many fewer transcribed regions ("seqfrags") outside known exons and ncRNAs. Most nonexonic seqfrags are in introns, raising the possibility that they are fragments of pre-mRNAs. The chromosomal locations of the majority of intergenic seqfrags in RNA-Seq data are near known genes, consistent with alternative cleavage and polyadenylation site usage, promoter- and terminator-associated transcripts, or new alternative exons; indeed, reads that bridge splice sites identified 4,544 new exons, affecting 3,554 genes. Most of the remaining seqfrags correspond to either single reads that display characteristics of random sampling from a low-level background or several thousand small transcripts (median length = 111 bp) present at higher levels, which also tend to display sequence conservation and originate from regions with open chromatin. We conclude that, while there are bona fide new intergenic transcripts, their number and abundance is generally low in comparison to known exons, and the genome is not as pervasively transcribed as previously reported.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
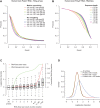
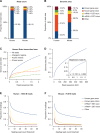
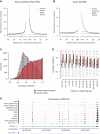
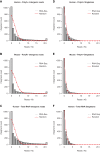

Comment in
-
Dark matter transcripts: sound and fury, signifying nothing?PLoS Biol. 2010 May 18;8(5):e1000370. doi: 10.1371/journal.pbio.1000370. PLoS Biol. 2010. PMID: 20502697 Free PMC article. No abstract available.
-
Transcriptomics: Throwing light on dark matter.Nat Rev Genet. 2010 Jul;11(7):455. doi: 10.1038/nrg2819. Nat Rev Genet. 2010. PMID: 20559328 No abstract available.
-
How to find TUFs.Nat Methods. 2010 Aug;7(8):582. doi: 10.1038/nmeth0810-582. Nat Methods. 2010. PMID: 20704018 No abstract available.
References
-
- Kapranov P, Cawley S. E, Drenkow J, Bekiranov S, Strausberg R. L, et al. Large-scale transcriptional activity in chromosomes 21 and 22. Science. 2002;296:916–919. - PubMed
-
- Cheng J, Kapranov P, Drenkow J, Dike S, Brubaker S, et al. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science. 2005;308:1149–1154. - PubMed
-
- Kapranov P, Cheng J, Dike S, Nix D. A, Duttagupta R, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

