HTS-Compatible Patient-Derived Cell-Based Assay to Identify Small Molecule Modulators of Aberrant Splicing in Myotonic Dystrophy Type 1
- PMID: 20502647
- PMCID: PMC2874217
- DOI: 10.2174/1875397301004010009
HTS-Compatible Patient-Derived Cell-Based Assay to Identify Small Molecule Modulators of Aberrant Splicing in Myotonic Dystrophy Type 1
Abstract
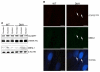
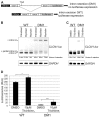
Myotonic dystrophy type 1 (DM1) is a genetic disorder characterized by muscle wasting, myotonia, cataracts, cardiac arrhythmia, hyperinsulinism and intellectual deficits, and is caused by expansion of a CTG repeat in the 3'UTR of the Dystrophia Myotonica-Protein Kinase (DMPK) gene. The DMPK transcripts containing expanded CUG repeats accumulate in nuclear foci and ultimately cause mis-splicing of secondary genes through the dysregulation of RNA-binding proteins including Muscleblind 1 (MBNL1) and CUG binding protein 1 (CUGBP1). Correction of mis-splicing of genes such as the Skeletal muscle-specific chloride channel 1 (CLCN1), Cardiac troponin T (TNNT2), Insulin receptor (INSR) and Sarcoplasmic/endoplasmic reticulum Ca(2+)ATPase 1 (SERCA1) may alleviate some of the symptoms of DM1; hence identification of small molecule modulators is an important step towards a therapy for DM1 patients. Here we describe the generation of immortalized myoblast cell lines derived from healthy (DMPK CTG(5)) and DM1 patient (DMPK CTG(1000)) fibroblasts by constitutive overexpression of human telomerase reverse transcriptase (hTERT) and inducible overexpression of the Myoblast determination factor (MYOD). MBNL1-containing nuclear foci, mis-splicing events and defective myotube differentiation defects characteristic of DM1 were observed in these cells. A CLCN1 luciferase minigene construct (CLCN1-luc) was stably introduced to monitor intron 2 retention in the DM1 cellular context (a reported splicing defect in DM1). The assay was validated by performing a high-throughput screen (HTS) of ~13,000 low molecular weight compounds against the CLCN1-luc DM1 myoblast cell line, providing an ideal system for conducting HTS to better understand and treat DM1.
Keywords: Alternative splicing; DM1; HTS; cell-based assay.; myotonic dystrophy.
Figures



Similar articles
-
Molecular Effects of the CTG Repeats in Mutant Dystrophia Myotonica Protein Kinase Gene.Curr Genomics. 2008 Dec;9(8):509-16. doi: 10.2174/138920208786847944. Curr Genomics. 2008. PMID: 19516957 Free PMC article.
-
Vorinostat Improves Myotonic Dystrophy Type 1 Splicing Abnormalities in DM1 Muscle Cell Lines and Skeletal Muscle from a DM1 Mouse Model.Int J Mol Sci. 2023 Feb 14;24(4):3794. doi: 10.3390/ijms24043794. Int J Mol Sci. 2023. PMID: 36835205 Free PMC article.
-
The CTG repeat expansion size correlates with the splicing defects observed in muscles from myotonic dystrophy type 1 patients.J Med Genet. 2008 Oct;45(10):639-46. doi: 10.1136/jmg.2008.058909. Epub 2008 Jul 8. J Med Genet. 2008. PMID: 18611984
-
An Overview of Alternative Splicing Defects Implicated in Myotonic Dystrophy Type I.Genes (Basel). 2020 Sep 22;11(9):1109. doi: 10.3390/genes11091109. Genes (Basel). 2020. PMID: 32971903 Free PMC article. Review.
-
Small Molecules Which Improve Pathogenesis of Myotonic Dystrophy Type 1.Front Neurol. 2018 May 18;9:349. doi: 10.3389/fneur.2018.00349. eCollection 2018. Front Neurol. 2018. PMID: 29867749 Free PMC article. Review.
Cited by
-
Pluripotent Stem Cells in Disease Modeling and Drug Discovery for Myotonic Dystrophy Type 1.Cells. 2023 Feb 10;12(4):571. doi: 10.3390/cells12040571. Cells. 2023. PMID: 36831237 Free PMC article. Review.
-
Promising AAV.U7snRNAs vectors targeting DMPK improve DM1 hallmarks in patient-derived cell lines.Front Cell Dev Biol. 2023 Jun 15;11:1181040. doi: 10.3389/fcell.2023.1181040. eCollection 2023. Front Cell Dev Biol. 2023. PMID: 37397246 Free PMC article.
-
Mitigating RNA Toxicity in Myotonic Dystrophy using Small Molecules.Int J Mol Sci. 2019 Aug 17;20(16):4017. doi: 10.3390/ijms20164017. Int J Mol Sci. 2019. PMID: 31426500 Free PMC article. Review.
-
Development of an AP-FRET based analysis for characterizing RNA-protein interactions in myotonic dystrophy (DM1).PLoS One. 2014 Apr 29;9(4):e95957. doi: 10.1371/journal.pone.0095957. eCollection 2014. PLoS One. 2014. PMID: 24781112 Free PMC article.
-
Myoblasts generated by lentiviral mediated MyoD transduction of myotonic dystrophy type 1 (DM1) fibroblasts can be used for assays of therapeutic molecules.BMC Res Notes. 2011 Nov 11;4:490. doi: 10.1186/1756-0500-4-490. BMC Res Notes. 2011. PMID: 22078098 Free PMC article.
References
-
- Machuca-Tzili L, Brook D, Hilton-Jones D. Clinical and molecular aspects of the myotonic dystrophies: a review. Muscle Nerve. 2005;32:1–18. - PubMed
-
- Mahadevan M, Tsilfidis C, Sabourin L, et al. Myotonic dystrophy mutation: an unstable CTG repeat in the 3' untranslated region of the gene. Science. 1992;255:1253–5. - PubMed
-
- Brook JD, McCurrach ME, Harley HG, et al. Molecular basis of myotonic dystrophy: expansion of a trinucleotide (CTG) repeat at the 3' end of a transcript encoding a protein kinase family member. Cell. 1992;69:385. - PubMed
-
- Gharehbaghi-Schnell EB, Finsterer J, Korschineck I, Mamoli B, Binder BR. Genotype-phenotype correlation in myotonic dystrophy. Clin Genet. 1998;53:20–6. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
