FACT prevents the accumulation of free histones evicted from transcribed chromatin and a subsequent cell cycle delay in G1
- PMID: 20502685
- PMCID: PMC2873916
- DOI: 10.1371/journal.pgen.1000964
FACT prevents the accumulation of free histones evicted from transcribed chromatin and a subsequent cell cycle delay in G1
Abstract
The FACT complex participates in chromatin assembly and disassembly during transcription elongation. The yeast mutants affected in the SPT16 gene, which encodes one of the FACT subunits, alter the expression of G1 cyclins and exhibit defects in the G1/S transition. Here we show that the dysfunction of chromatin reassembly factors, like FACT or Spt6, down-regulates the expression of the gene encoding the cyclin that modulates the G1 length (CLN3) in START by specifically triggering the repression of its promoter. The G1 delay undergone by spt16 mutants is not mediated by the DNA-damage checkpoint, although the mutation of RAD53, which is otherwise involved in histone degradation, enhances the cell-cycle defects of spt16-197. We reveal how FACT dysfunction triggers an accumulation of free histones evicted from transcribed chromatin. This accumulation is enhanced in a rad53 background and leads to a delay in G1. Consistently, we show that the overexpression of histones in wild-type cells down-regulates CLN3 in START and causes a delay in G1. Our work shows that chromatin reassembly factors are essential players in controlling the free histones potentially released from transcribed chromatin and describes a new cell cycle phenomenon that allows cells to respond to excess histones before starting DNA replication.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
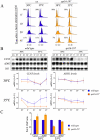


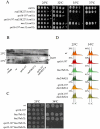



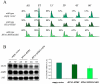
Similar articles
-
A gene-specific requirement for FACT during transcription is related to the chromatin organization of the transcribed region.Mol Cell Biol. 2006 Dec;26(23):8710-21. doi: 10.1128/MCB.01129-06. Epub 2006 Sep 25. Mol Cell Biol. 2006. PMID: 17000768 Free PMC article.
-
FACT, the Bur kinase pathway, and the histone co-repressor HirC have overlapping nucleosome-related roles in yeast transcription elongation.PLoS One. 2011;6(10):e25644. doi: 10.1371/journal.pone.0025644. Epub 2011 Oct 12. PLoS One. 2011. PMID: 22022426 Free PMC article.
-
An Intrinsically Disordered Region of the FACT Subunit, Spt16, Promotes Chromatin Disassembly in Stimulating the Pre-Initiation Complex Formation at the Promoter for Transcription Initiation In Vivo.Mol Cell Biol. 2025;45(7):263-282. doi: 10.1080/10985549.2025.2501630. Epub 2025 May 23. Mol Cell Biol. 2025. PMID: 40405832
-
Facts about FACT and transcript elongation through chromatin.Curr Opin Genet Dev. 2004 Apr;14(2):139-46. doi: 10.1016/j.gde.2004.02.004. Curr Opin Genet Dev. 2004. PMID: 15196460 Review.
-
Opposing Roles of FACT for Euchromatin and Heterochromatin in Yeast.Biomolecules. 2023 Feb 16;13(2):377. doi: 10.3390/biom13020377. Biomolecules. 2023. PMID: 36830746 Free PMC article. Review.
Cited by
-
The Histone Chaperone NRP1 Interacts with WEREWOLF to Activate GLABRA2 in Arabidopsis Root Hair Development.Plant Cell. 2017 Feb;29(2):260-276. doi: 10.1105/tpc.16.00719. Epub 2017 Jan 30. Plant Cell. 2017. PMID: 28138017 Free PMC article.
-
Defective transfer of parental histone decreases frequency of homologous recombination by increasing free histone pools in budding yeast.Nucleic Acids Res. 2024 May 22;52(9):5138-5151. doi: 10.1093/nar/gkae205. Nucleic Acids Res. 2024. PMID: 38554108 Free PMC article.
-
Asymmetric Transcription Factor Partitioning During Yeast Cell Division Requires the FACT Chromatin Remodeler and Cell Cycle Progression.Genetics. 2020 Nov;216(3):701-716. doi: 10.1534/genetics.120.303439. Epub 2020 Sep 2. Genetics. 2020. PMID: 32878900 Free PMC article.
-
Uncoupling histone turnover from transcription-associated histone H3 modifications.Nucleic Acids Res. 2015 Apr 30;43(8):3972-85. doi: 10.1093/nar/gkv282. Epub 2015 Apr 6. Nucleic Acids Res. 2015. PMID: 25845593 Free PMC article.
-
Extra-nuclear histones: origin, significance and perspectives.Mol Cell Biochem. 2022 Feb;477(2):507-524. doi: 10.1007/s11010-021-04300-4. Epub 2021 Nov 18. Mol Cell Biochem. 2022. PMID: 34796445 Review.
References
-
- Reinberg D, Sims RJ., 3rd de FACTo nucleosome dynamics. J Biol Chem. 2006;281:23297–23301. - PubMed
-
- Formosa T. FACT and the reorganized nucleosome. Mol Biosyst. 2008;4:1085–1093. - PubMed
-
- Orphanides G, LeRoy G, Chang CH, Luse DS, Reinberg D. FACT, a factor that facilitates transcript elongation through nucleosomes. Cell. 1998;92:105–116. - PubMed
-
- Pavri R, Zhu B, Li G, Trojer P, Mandal S, et al. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell. 2006;125:703–717. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

