The origin and genetic variation of domestic chickens with special reference to junglefowls Gallus g. gallus and G. varius
- PMID: 20502703
- PMCID: PMC2873279
- DOI: 10.1371/journal.pone.0010639
The origin and genetic variation of domestic chickens with special reference to junglefowls Gallus g. gallus and G. varius
Abstract
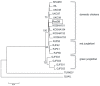
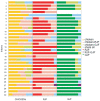
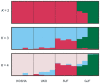
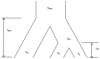
It is postulated that chickens (Gallus gallus domesticus) became domesticated from wild junglefowls in Southeast Asia nearly 10,000 years ago. Based on 19 individual samples covering various chicken breeds, red junglefowl (G. g. gallus), and green junglefowl (G. varius), we address the origin of domestic chickens, the relative roles of ancestral polymorphisms and introgression, and the effects of artificial selection on the domestic chicken genome. DNA sequences from 30 introns at 25 nuclear loci are determined for both diploid chromosomes from a majority of samples. The phylogenetic analysis shows that the DNA sequences of chickens, red and green junglefowls formed reciprocally monophyletic clusters. The Markov chain Monte Carlo simulation further reveals that domestic chickens diverged from red junglefowl 58,000+/-16,000 years ago, well before the archeological dating of domestication, and that their common ancestor in turn diverged from green junglefowl 3.6 million years ago. Several shared haplotypes nonetheless found between green junglefowl and chickens are attributed to recent unidirectional introgression of chickens into green junglefowl. Shared haplotypes are more frequently found between red junglefowl and chickens, which are attributed to both introgression and ancestral polymorphisms. Within each chicken breed, there is an excess of homozygosity, but there is no significant reduction in the nucleotide diversity. Phenotypic modifications of chicken breeds as a result of artificial selection appear to stem from ancestral polymorphisms at a limited number of genetic loci.
Conflict of interest statement
Figures

Similar articles
-
Origin and evolutionary history of domestic chickens inferred from a large population study of Thai red junglefowl and indigenous chickens.Sci Rep. 2021 Jan 21;11(1):2035. doi: 10.1038/s41598-021-81589-7. Sci Rep. 2021. PMID: 33479400 Free PMC article.
-
The wild species genome ancestry of domestic chickens.BMC Biol. 2020 Feb 12;18(1):13. doi: 10.1186/s12915-020-0738-1. BMC Biol. 2020. PMID: 32050971 Free PMC article.
-
The origin and genetic diversity of Chinese native chicken breeds.Biochem Genet. 2002 Jun;40(5-6):163-74. doi: 10.1023/a:1015832108669. Biochem Genet. 2002. PMID: 12137331
-
Chicken domestication: from archeology to genomics.C R Biol. 2011 Mar;334(3):197-204. doi: 10.1016/j.crvi.2010.12.012. C R Biol. 2011. PMID: 21377614 Review.
-
Domestic chicken diversity: Origin, distribution, and adaptation.Anim Genet. 2021 Aug;52(4):385-394. doi: 10.1111/age.13091. Epub 2021 May 31. Anim Genet. 2021. PMID: 34060099 Review.
Cited by
-
Early Holocene chicken domestication in northern China.Proc Natl Acad Sci U S A. 2014 Dec 9;111(49):17564-9. doi: 10.1073/pnas.1411882111. Epub 2014 Nov 24. Proc Natl Acad Sci U S A. 2014. PMID: 25422439 Free PMC article.
-
Thinking chickens: a review of cognition, emotion, and behavior in the domestic chicken.Anim Cogn. 2017 Mar;20(2):127-147. doi: 10.1007/s10071-016-1064-4. Epub 2017 Jan 2. Anim Cogn. 2017. PMID: 28044197 Free PMC article. Review.
-
Biophysics and population size constrains speciation in an evolutionary model of developmental system drift.PLoS Comput Biol. 2019 Jul 23;15(7):e1007177. doi: 10.1371/journal.pcbi.1007177. eCollection 2019 Jul. PLoS Comput Biol. 2019. PMID: 31335870 Free PMC article.
-
Deeper insight into maternal genetic assessments and demographic history for Egyptian indigenous chicken populations using mtDNA analysis.J Adv Res. 2016 Sep;7(5):615-23. doi: 10.1016/j.jare.2016.06.005. Epub 2016 Jun 28. J Adv Res. 2016. PMID: 27489728 Free PMC article.
-
A genome-wide scan to identify signatures of selection in two Iranian indigenous chicken ecotypes.Genet Sel Evol. 2021 Sep 9;53(1):72. doi: 10.1186/s12711-021-00664-9. Genet Sel Evol. 2021. PMID: 34503452 Free PMC article.
References
-
- Sutter NB, Bustamante CD, Chase K, Gray MM, Zhao K, et al. A single IGF1 allele is a major determinant of small size in dogs. Science. 2007;316:112–115. DOI: 10.1126/science.1137045. - DOI - PMC - PubMed
-
- Anderson TM, vonHoldt BM, Candille SI, Musiani M, Greco C, et al. Molecular and evolutionary history of melanism in North American gray wolves. Science. 2009;323:1339–1343. DOI: 10.1126/science.1165448. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

