The capsid protein of Turnip crinkle virus overcomes two separate defense barriers to facilitate systemic movement of the virus in Arabidopsis
- PMID: 20504923
- PMCID: PMC2897622
- DOI: 10.1128/JVI.02643-09
The capsid protein of Turnip crinkle virus overcomes two separate defense barriers to facilitate systemic movement of the virus in Arabidopsis
Abstract
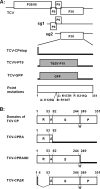
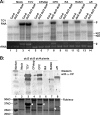
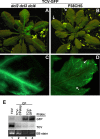
The capsid protein (CP) of Turnip crinkle virus (TCV) is a multifunctional protein needed for virus assembly, suppression of RNA silencing-based antiviral defense, and long-distance movement in infected plants. In this report, we have examined genetic requirements for the different functions of TCV CP and evaluated the interdependence of these functions. A series of TCV mutants containing alterations in the CP coding region were generated. These alterations range from single-amino-acid substitutions and domain truncations to knockouts of CP translation. The latter category also contained two constructs in which the CP coding region was replaced by either the cDNA of a silencing suppressor of a different virus or that of green fluorescent protein. These mutants were used to infect Arabidopsis plants with diminished antiviral silencing capability (dcl2 dcl3 dcl4 plants). There was a strong correlation between the ability of mutants to reach systemic leaves and the silencing suppressor activity of mutant CP. Virus particles were not essential for entry of the viral genome into vascular bundles in the inoculated leaves in the absence of antiviral silencing. However, virus particles were necessary for egress of the viral genome from the vasculature of systemic leaves. Our experiments demonstrate that TCV CP not only allows the viral genome to access the systemic movement channel through silencing suppression but also ensures its smooth egress by way of assembled virus particles. These results illustrate that efficient long-distance movement of TCV requires both functions afforded by the CP.
Figures


References
-
- Carrington, J. C., T. J. Morris, P. G. Stockley, and S. C. Harrison. 1987. Structure and assembly of turnip crinkle virus. IV. Analysis of the coat protein gene and implications of the subunit primary structure. J. Mol. Biol. 194:265-276. - PubMed
-
- Chen, M.-H., and V. Citovsky. 2003. Systemic movement of a tobamovirus requires host cell pectin methylesterase. Plant J. 35:386-392. - PubMed
-
- Choi, C. W., F. Qu, T. Ren, X. Ye, and T. J. Morris. 2004. The RNA silencing suppressor function of Turnip crinkle virus coat protein cannot be attributed to its interaction with the Arabidopsis protein TIP. J. Gen. Virol. 85:3415-3420. - PubMed
-
- Cohen, Y., A. Gisel, and P. C. Zambryski. 2000. Cell-to-cell and systemic movement of recombinant green fluorescent protein-tagged turnip crinkle viruses. Virology 273:258-266. - PubMed
-
- Deleris, A., J. Gallego-Bartolome, J. Bao, K. D. Kasschau, J. C. Carrington, and O. Voinnet. 2006. Hierarchical action and inhibition of plant Dicer-like proteins in antiviral defense. Science 313:68-71. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

