Subunits of the Drosophila CCR4-NOT complex and their roles in mRNA deadenylation
- PMID: 20504953
- PMCID: PMC2885685
- DOI: 10.1261/rna.2145110
Subunits of the Drosophila CCR4-NOT complex and their roles in mRNA deadenylation
Abstract
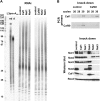
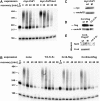
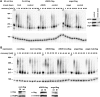
The CCR4-NOT complex is the main enzyme catalyzing the deadenylation of mRNA. We have investigated the composition of this complex in Drosophila melanogaster by immunoprecipitation with a monoclonal antibody directed against NOT1. The CCR4, CAF1 (=POP2), NOT1, NOT2, NOT3, and CAF40 subunits were associated in a stable complex, but NOT4 was not. Factors known to be involved in mRNA regulation were prominent among the other proteins coprecipitated with the CCR4-NOT complex, as analyzed by mass spectrometry. The complex was localized mostly in the cytoplasm but did not appear to be a major component of P bodies. Of the known CCR4 paralogs, Nocturnin was found associated with the subunits of the CCR4-NOT complex, whereas Angel and 3635 were not. RNAi experiments in Schneider cells showed that CAF1, NOT1, NOT2, and NOT3 are required for bulk poly(A) shortening and hsp70 mRNA deadenylation, but knock-down of CCR4, CAF40, and NOT4 did not affect these processes. Overexpression of catalytically dead CAF1 had a dominant-negative effect on mRNA decay. In contrast, overexpression of inactive CCR4 had no effect. We conclude that CAF1 is the major catalytically important subunit of the CCR4-NOT complex in Drosophila Schneider cells. Nocturnin may also be involved in mRNA deadenylation, whereas there is no evidence for a similar role of Angel and 3635.
Figures





Similar articles
-
A complex containing the CCR4 and CAF1 proteins is involved in mRNA deadenylation in Drosophila.EMBO J. 2004 Jul 21;23(14):2862-71. doi: 10.1038/sj.emboj.7600273. Epub 2004 Jun 24. EMBO J. 2004. PMID: 15215893 Free PMC article.
-
NOT10 and C2orf29/NOT11 form a conserved module of the CCR4-NOT complex that docks onto the NOT1 N-terminal domain.RNA Biol. 2013 Feb;10(2):228-44. doi: 10.4161/rna.23018. Epub 2013 Jan 9. RNA Biol. 2013. PMID: 23303381 Free PMC article.
-
The structural basis for the interaction between the CAF1 nuclease and the NOT1 scaffold of the human CCR4-NOT deadenylase complex.Nucleic Acids Res. 2012 Nov;40(21):11058-72. doi: 10.1093/nar/gks883. Epub 2012 Sep 12. Nucleic Acids Res. 2012. PMID: 22977175 Free PMC article.
-
The Ccr4-Not Complex: Architecture and Structural Insights.Subcell Biochem. 2017;83:349-379. doi: 10.1007/978-3-319-46503-6_13. Subcell Biochem. 2017. PMID: 28271483 Review.
-
Heterogeneity and complexity within the nuclease module of the Ccr4-Not complex.Front Genet. 2013 Dec 23;4:296. doi: 10.3389/fgene.2013.00296. Front Genet. 2013. PMID: 24391663 Free PMC article. Review.
Cited by
-
Sexual and asexual oogenesis require the expression of unique and shared sets of genes in the insect Acyrthosiphon pisum.BMC Genomics. 2012 Feb 15;13:76. doi: 10.1186/1471-2164-13-76. BMC Genomics. 2012. PMID: 22336141 Free PMC article.
-
The Not4 E3 ligase and CCR4 deadenylase play distinct roles in protein quality control.PLoS One. 2014 Jan 17;9(1):e86218. doi: 10.1371/journal.pone.0086218. eCollection 2014. PLoS One. 2014. PMID: 24465968 Free PMC article.
-
Regulatory principles governing the maternal-to-zygotic transition: insights from Drosophila melanogaster.Open Biol. 2018 Dec;8(12):180183. doi: 10.1098/rsob.180183. Open Biol. 2018. PMID: 30977698 Free PMC article. Review.
-
Divergence of the expression and subcellular localization of CCR4-associated factor 1 (CAF1) deadenylase proteins in Oryza sativa.Plant Mol Biol. 2014 Jul;85(4-5):443-58. doi: 10.1007/s11103-014-0196-7. Epub 2014 May 8. Plant Mol Biol. 2014. PMID: 24805883
-
The Regulatory Properties of the Ccr4-Not Complex.Cells. 2020 Oct 29;9(11):2379. doi: 10.3390/cells9112379. Cells. 2020. PMID: 33138308 Free PMC article. Review.
References
-
- Aviv T, Lin Z, Lau S, Rendl LM, Sicheri F, Smibert CA 2003. The RNA binding SAM domain of Smaug defines a new family of post-transcriptional regulators. Nat Struct Biol 10: 614–621 - PubMed
-
- Baggs JE, Green CB 2003. Nocturnin, a deadenylase in Xenopus laevis retina: A mechanism for posttranscriptional control of circadian-related mRNA. Curr Biol 13: 189–198 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
