Evaluation of the analytical performance of the Xpert MTB/RIF assay
- PMID: 20504986
- PMCID: PMC2897495
- DOI: 10.1128/JCM.00128-10
Evaluation of the analytical performance of the Xpert MTB/RIF assay
Abstract
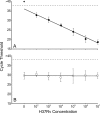
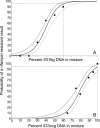
We performed the first studies of analytic sensitivity, analytic specificity, and dynamic range for the new Xpert MTB/RIF assay, a nucleic acid amplification-based diagnostic system that detects Mycobacterium tuberculosis and rifampin (RIF) resistance in under 2 h. The sensitivity of the assay was tested with 79 phylogenetically and geographically diverse M. tuberculosis isolates, including 42 drug-susceptible isolates and 37 RIF-resistant isolates containing 13 different rpoB mutations or mutation combinations. The specificity of the assay was tested with 89 nontuberculosis bacteria, fungi, and viruses. The Xpert MTB/RIF assay correctly identified all 79 M. tuberculosis isolates and correctly excluded all 89 nontuberculosis isolates. RIF resistance was correctly identified in all 37 resistant isolates and in none of the 42 susceptible isolates. Dynamic range was assessed by adding 10(2) to 10(7) CFU of M. tuberculosis into M. tuberculosis-negative sputum samples. The assay showed a log-linear relationship between cycle threshold and input CFU over the entire concentration range. Resistance detection in the presence of different mixtures of RIF-resistant and RIF-susceptible DNA was assessed. Resistance detection was dependent on the particular mutation and required between 65% and 100% mutant DNA to be present in the sample for 95% certainty of resistance detection. Finally, we studied whether assay specificity could be affected by cross-contaminating amplicons generated by the GenoType MTBDRplus assay. M. tuberculosis was not detected until at least 10(8) copies of an MTBDRplus amplicon were spiked into 1 ml of sputum, suggesting that false-positive results would be unlikely to occur.
Figures

References
-
- Balasingham, S. V., T. Davidsen, I. Szpinda, S. A. Frye, and T. Tonjum. 2009. Molecular diagnostics in tuberculosis: basis and implications for therapy. Mol. Diagn. Ther. 13:137-151. - PubMed
-
- Chakravorty, S., B. Aladegbami, M. Burday, M. Levi, S. A. Marras, D. Shah, H. H. El-Hajj, F. R. Kramer, and D. Alland. 2010. Rapid universal identification of bacterial pathogens from clinical cultures using a novel sloppy molecular beacon melting temperature signature technique. J. Clin. Microbiol. 48:258-267. - PMC - PubMed
-
- Chakravorty, S., B. Aladegbami, A. S. Motiwala, Y. Dai, H. Safi, M. Brimacombe, D. Helb, and D. Alland. 2008. Rifampin resistance, Beijing-W clade-single nucleotide polymorphism cluster group 2 phylogeny, and the Rv2629 191-C allele in Mycobacterium tuberculosis strains. J. Clin. Microbiol. 46:2555-2560. - PMC - PubMed
-
- Colangeli, R., D. Helb, C. Vilcheze, M. H. Hazbon, C. G. Lee, H. Safi, B. Sayers, I. Sardone, M. B. Jones, R. D. Fleischmann, S. N. Peterson, W. R. Jacobs, Jr., and D. Alland. 2007. Transcriptional regulation of multi-drug tolerance and antibiotic-induced responses by the histone-like protein Lsr2 in M. tuberculosis. PLoS Pathog. 3:e87. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

