Mitochondrial DNA damage as a potential mechanism for age-related macular degeneration
- PMID: 20505194
- PMCID: PMC3061495
- DOI: 10.1167/iovs.10-5429
Mitochondrial DNA damage as a potential mechanism for age-related macular degeneration
Abstract
Purpose: Increasing evidence suggests a central role for mitochondrial (mt) dysfunction in age-related macular degeneration (AMD). Previous proteomic data from the retinal pigment epithelium (RPE) revealed significant changes to mt proteins, suggesting potential functional defects and damage to mitochondrial DNA (mtDNA) with AMD progression. The present study tests the hypothesis that mtDNA damage increases with aging and AMD.
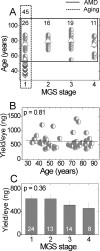
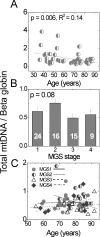
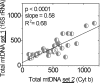
Methods: Genomic DNA was isolated from the macular region of human donor RPE graded for stages of AMD (Minnesota Grading System [MGS] 1-4). Region-specific mtDNA damage with normal aging was evaluated in 45 control subjects (ages 34-88 years, MGS 1) and AMD-associated damage in diseased subjects (n = 46), compared with that in age-matched control subjects (n = 26). Lesions per 10 kb per genome in the mtDNA and nuclear DNA were measured with long-extension polymerase chain reaction (LX PCR). The level of deleted mtDNA in each donor was measured with quantitative real-time PCR (qPCR).
Results: With aging, an increase in mtDNA damage was observed only in the common deletion region of the mt genome. In contrast, with AMD, mtDNA lesions increased significantly in all regions of the mt genome beyond levels found in age-matched control subjects. mtDNA accumulated more lesions than did two nuclear genes, with total damage of the mt genome estimated to be eight times higher.
Conclusions: Collectively, the data indicate that mtDNA is preferentially damaged with AMD progression. These results suggest a potential link between mt dysfunction due to increased mtDNA lesions and AMD.
Figures




References
-
- Friedman DS, O'Colmain BJ, Munoz B, et al. Prevalence of age-related macular degeneration in the United States. Arch Ophthalmol. 2004;122:564–572 - PubMed
-
- Brown MM, Brown GC, Sharma S, et al. The burden of age-related macular degeneration: a value-based analysis. Curr Opin Ophthalmol. 2006;17:257–266 - PubMed
-
- Brown DM, Kaiser PK, Michels M, et al. Ranibizumab versus verteporfin for neovascular age-related macular degeneration. N Engl J Med. 2006;355:1432–1444 - PubMed
-
- Rosenfeld PJ, Brown DM, Heier JS, et al. Ranibizumab for neovascular age-related macular degeneration. N Engl J Med. 2006;355:1419–1431 - PubMed
-
- Strauss O. The retinal pigment epithelium in visual function. Physiol Rev. 2005;85:845–881 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

