microRNA expression patterns reveal differential expression of target genes with age
- PMID: 20505758
- PMCID: PMC2873959
- DOI: 10.1371/journal.pone.0010724
microRNA expression patterns reveal differential expression of target genes with age
Abstract
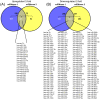
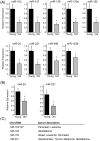
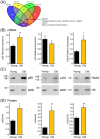
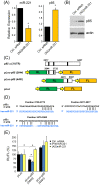
Recent evidence supports a role for microRNAs (miRNAs) in regulating the life span of model organisms. However, little is known about how these small RNAs contribute to human aging. Here, we profiled the expression of over 800 miRNAs in peripheral blood mononuclear cells from young and old individuals by real-time RT-PCR analysis. This genome-wide assessment of miRNA expression revealed that the majority of miRNAs studied decreased in abundance with age. We identified nine miRNAs (miR-103, miR-107, miR-128, miR-130a, miR-155, miR-24, miR-221, miR-496, miR-1538) that were significantly lower in older individuals. Among them, five have been implicated in cancer pathogenesis. Predicted targets of several of these miRNAs, including PI3 kinase (PI3K), c-Kit and H2AX, were found to be elevated with advancing age, supporting a possible role for them in the aging process. Furthermore, we found that decreasing the levels of miR-221 was sufficient to cause a corresponding increase in the expression of the predicted target, PI3K. Taken together, these findings demonstrate that changes in miRNA expression occur with human aging and suggest that miRNAs and their predicted targets have the potential to be diagnostic indicators of age or age-related diseases.
Conflict of interest statement
Figures

References
-
- Chekulaeva M, Filipowicz W. Mechanisms of miRNA-mediated post-transcriptional regulation in animal cells. Curr Opin Cell Biol. 2009;21:452–460. - PubMed
-
- Garzon R, Calin GA, Croce CM. MicroRNAs in Cancer. Annu Rev Med. 2009;60:167–179. - PubMed
-
- Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–139. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

