Cartography of methicillin-resistant S. aureus transcripts: detection, orientation and temporal expression during growth phase and stress conditions
- PMID: 20505759
- PMCID: PMC2873960
- DOI: 10.1371/journal.pone.0010725
Cartography of methicillin-resistant S. aureus transcripts: detection, orientation and temporal expression during growth phase and stress conditions
Abstract
Background: Staphylococcus aureus is a versatile bacterial opportunist responsible for a wide spectrum of infections. The severity of these infections is highly variable and depends on multiple parameters including the genome content of the bacterium as well as the condition of the infected host. Clinically and epidemiologically, S. aureus shows a particular capacity to survive and adapt to drastic environmental changes including the presence of numerous antimicrobial agents. Mechanisms triggering this adaptation remain largely unknown despite important research efforts. Most studies evaluating gene content have so far neglected to analyze the so-called intergenic regions as well as potential antisense RNA molecules.
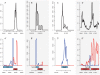
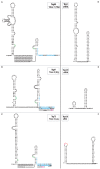
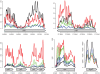
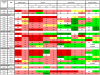
Principal findings: Using high-throughput sequencing technology, we performed an inventory of the whole transcriptome of S. aureus strain N315. In addition to the annotated transcription units, we identified more than 195 small transcribed regions, in the chromosome and the plasmid of S. aureus strain N315. The coding strand of each transcript was identified and structural analysis enabled classification of all discovered transcripts. RNA purified at four time-points during the growth phase of the bacterium allowed us to define the temporal expression of such transcripts. A selection of 26 transcripts of interest dispersed along the intergenic regions was assessed for expression changes in the presence of various stress conditions including pH, temperature, oxidative shocks and growth in a stringent medium. Most of these transcripts showed expression patterns specific for the defined stress conditions that we tested.
Conclusions: These RNA molecules potentially represent important effectors of S. aureus adaptation and more generally could support some of the epidemiological characteristics of the bacterium.
Conflict of interest statement
Figures

References
-
- Boyce JM. Methicillin-resistant Staphylococcus aureus in hospitals and long-term care facilities: microbiology, epidemiology, and preventive measures. Infect Control Hosp Epidemiol. 1992;13:725–737. - PubMed
-
- Kazakova SV, Hageman JC, Matava M, Srinivasan A, Phelan L, et al. A clone of methicillin-resistant Staphylococcus aureus among professional football players. N Engl J Med. 2005;352:468–475. - PubMed
-
- Waldvogel FA, Bisno AL. Infections associated with indwelling medical devices. Washington, DC: ASM Press; 2000. 436
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

