In vivo identification of solar radiation-responsive gene network: role of the p38 stress-dependent kinase
- PMID: 20505830
- PMCID: PMC2874014
- DOI: 10.1371/journal.pone.0010776
In vivo identification of solar radiation-responsive gene network: role of the p38 stress-dependent kinase
Abstract
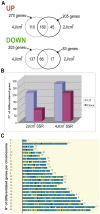
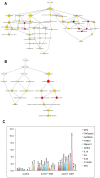
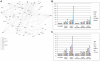
Solar radiation is one of the most common threats to the skin, with exposure eliciting a specific protective cellular response. To decrypt the underlying mechanism, we used whole genome microarrays (Agilent 44K) to study epidermis gene expression in vivo in skin exposed to simulated solar radiation (SSR). We procured epidermis samples from healthy Caucasian patients, with phototypes II or III, and used two different SSR doses (2 and 4 J/cm(2)), the lower of which corresponded to the minimal erythemal dose. Analyses were carried out five hours after irradiation to identify early gene expression events in the photoprotective response. About 1.5% of genes from the human genome showed significant changes in gene expression. The annotations of these affected genes were assessed. They indicated a strengthening of the inflammation process and up-regulation of the JAK-STAT pathway and other pathways. Parallel to the p53 pathway, the p38 stress-responsive pathway was affected, supporting and mediating p53 function. We used an ex vivo assay with a specific inhibitor of p38 (SB203580) to investigate genes the expression of which was associated with active p38 kinase. We identified new direct p38 target genes and further characterized the role of p38. Our findings provide further insight into the physiological response to UV, including cell-cell interactions and cross-talk effects.
Conflict of interest statement
Figures


Similar articles
-
Paeonol suppresses solar ultraviolet-induced skin inflammation by targeting T-LAK cell-originated protein kinase.Oncotarget. 2017 Apr 18;8(16):27093-27104. doi: 10.18632/oncotarget.15636. Oncotarget. 2017. PMID: 28404919 Free PMC article.
-
Cefradine blocks solar-ultraviolet induced skin inflammation through direct inhibition of T-LAK cell-originated protein kinase.Oncotarget. 2016 Apr 26;7(17):24633-45. doi: 10.18632/oncotarget.8260. Oncotarget. 2016. PMID: 27016423 Free PMC article.
-
Role of p38 MAPK in UVB-induced inflammatory responses in the skin of SKH-1 hairless mice.J Invest Dermatol. 2005 Jun;124(6):1318-25. doi: 10.1111/j.0022-202X.2005.23747.x. J Invest Dermatol. 2005. PMID: 15955110
-
Activation of mammalian gene expression by the UV component of sunlight--from models to reality.Bioessays. 1996 Feb;18(2):139-48. doi: 10.1002/bies.950180210. Bioessays. 1996. PMID: 8851047 Review.
-
p38 MAPK in regulating cellular responses to ultraviolet radiation.J Biomed Sci. 2007 May;14(3):303-12. doi: 10.1007/s11373-007-9148-4. Epub 2007 Mar 3. J Biomed Sci. 2007. PMID: 17334833 Review.
Cited by
-
Flipping the Switch: Innovations in Inducible Probes for Protein Profiling.ACS Chem Biol. 2021 Dec 17;16(12):2719-2730. doi: 10.1021/acschembio.1c00572. Epub 2021 Nov 15. ACS Chem Biol. 2021. PMID: 34779621 Free PMC article. Review.
-
Gene profiling of narrowband UVB-induced skin injury defines cellular and molecular innate immune responses.J Invest Dermatol. 2013 Mar;133(3):692-701. doi: 10.1038/jid.2012.359. Epub 2012 Nov 15. J Invest Dermatol. 2013. PMID: 23151847 Free PMC article.
-
Low dose ultraviolet B irradiation increases hyaluronan synthesis in epidermal keratinocytes via sequential induction of hyaluronan synthases Has1-3 mediated by p38 and Ca2+/calmodulin-dependent protein kinase II (CaMKII) signaling.J Biol Chem. 2013 Jun 21;288(25):17999-8012. doi: 10.1074/jbc.M113.472530. Epub 2013 May 3. J Biol Chem. 2013. PMID: 23645665 Free PMC article.
-
Possible Mechanisms of Oxidative Stress-Induced Skin Cellular Senescence, Inflammation, and Cancer and the Therapeutic Potential of Plant Polyphenols.Int J Mol Sci. 2023 Feb 13;24(4):3755. doi: 10.3390/ijms24043755. Int J Mol Sci. 2023. PMID: 36835162 Free PMC article. Review.
-
Merkel cell polyomavirus small T antigen mRNA level is increased following in vivo UV-radiation.PLoS One. 2010 Jul 2;5(7):e11423. doi: 10.1371/journal.pone.0011423. PLoS One. 2010. PMID: 20625394 Free PMC article. Clinical Trial.
References
-
- Ichihashi M, Ueda M, Budiyanto A, Bito T, Oka M, et al. UV-induced skin damage. Toxicology. 2003;189:21–39. - PubMed
-
- Thannickal VJ, Fanburg BL. Reactive oxygen species in cell signaling. Am J Physiol Lung Cell Mol Physiol. 2000;279:L1005–28. - PubMed
-
- Finkel T. Oxygen radicals and signaling. Curr Opin Cell Biol. 1998;10:248–53. - PubMed
-
- Rhee Sg. Redox signaling: hydrogen peroxide as intracellular messenger. Exp Mol Med. 1999;31:53–9. - PubMed
-
- Ravanat JL, Douki T, Cadet J. Direct and indirect effects of UV radiation on DNA and its components. J Photochem Photobiol B. 2001;63:88–102. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

