Gene optimization mechanisms: a multi-gene study reveals a high success rate of full-length human proteins expressed in Escherichia coli
- PMID: 20506237
- PMCID: PMC2970903
- DOI: 10.1002/pro.408
Gene optimization mechanisms: a multi-gene study reveals a high success rate of full-length human proteins expressed in Escherichia coli
Abstract
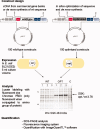
The genetic code is universal, but recombinant protein expression in heterologous systems is often hampered by divergent codon usage. Here, we demonstrate that reprogramming by standardized multi-parameter gene optimization software and de novo gene synthesis is a suitable general strategy to improve heterologous protein expression. This study compares expression levels of 94 full-length human wt and sequence-optimized genes coding for pharmaceutically important proteins such as kinases and membrane proteins in E. coli. Fluorescence-based quantification revealed increased protein yields for 70% of in vivo expressed optimized genes compared to the wt DNA sequences and also resulted in increased amounts of protein that can be purified. The improvement in transgene expression correlated with higher mRNA levels in our analyzed examples. In all cases tested, expression levels using wt genes in tRNA-supplemented bacterial strains were outperformed by optimized genes expressed in non-supplemented host cells.
Figures

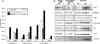

References
-
- Gräslund S, Nordlund P, Weigelt J, Hallberg BM, Bray J, Gileadi O, Knapp S, Oppermann U, Arrowsmith C, Hui R, Ming J, dhe-Paganon S, Park HW, Savchenko A, Yee A, Edwards A, Vincentelli R, Cambillau C, Kim R, Kim SH, Rao Z, Shi Y, Terwilliger TC, Kim CY, Hung LW, Waldo GS, Peleg Y, Albeck S, Unger T, Dym O, Prilusky J, Sussman JL, Stevens RC, Lesley SA, Wilson IA, Joachimiak A, Collart F, Dementieva I, Donnelly MI, Eschenfeldt WH, Kim Y, Stols L, Wu R, Zhou M, Burley SK, Emtage JS, Sauder JM, Thompson D, Bain K, Luz J, Gheyi T, Zhang F, Atwell S, Almo SC, Bonanno JB, Fiser A, Swaminathan S, Studier FW, Chance MR, Sali A, Acton TB, Xiao R, Zhao L, Ma LC, Hunt JF, Tong L, Cunningham K, Inouye M, Anderson S, Janjua H, Shastry R, Ho CK, Wang D, Wang H, Jiang M, Montelione GT, Stuart DI, Owens RJ, Daenke S, Schütz A, Heinemann U, Yokoyama S, Büssow K, Gunsalus KC. Protein production and purification. Nat Methods. 2008;5:135–146. - PMC - PubMed
-
- Swartz JR. Advances in Escherichia coli production of therapeutic proteins. Curr Opin Biotechnol. 2001;12:195–201. - PubMed
-
- Zhou Z, Schnake P, Xiao L, Lal AA. Enhanced expression of a recombinant malaria candidate vaccine in Escherichia coli by codon optimization. Protein Expr. Purif. 2004;34:87–94. - PubMed
-
- Nomura M, Ohsuye K, Mizuno A, Sakuragawa Y, Tanaka S. Influence of messenger RNA secondary structure on translation efficiency. Nucleic Acids Symp. Ser. 1984;15:173–176. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

