How sequence directs bending in tropomyosin and other two-stranded alpha-helical coiled coils
- PMID: 20506487
- PMCID: PMC2974828
- DOI: 10.1002/pro.415
How sequence directs bending in tropomyosin and other two-stranded alpha-helical coiled coils
Abstract
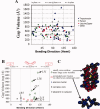
A quantitative analysis of the direction of bending of two-stranded alpha-helical coiled coils in crystal structures has been carried out to help determine how the amino acid sequence of the coiled coil influences its shape and function. Change in the axial staggering of the coiled coil, occurring at the boundaries of either clusters of core alanines in tropomyosin or of clusters of core bulky residues in the myosin rod, causes bending within the plane of the local dimer. The results also reveal that large gaps in the core of the coiled coil, which are seen for small core residues near large core residues or for unbranched core residues near canonical branched core residues, are correlated with bending out of the local dimeric plane. Comparison of tropomyosin structures determined in independent crystal environments provides further evidence for the concept that sequence directs the bending of the coiled coil, but that crystal environment is at least as important as sequence for determining the magnitude of bending. Tropomyosin thus appears to consist of more directionally restrained hinge-like joints rather than directionally variable universal joints, which helps account for and predicts the geometric and dynamic nature of its binding to F-actin.
Figures



References
-
- O'Shea EK, Klemm JD, Kim PS, Alber T. X-ray structure of the GCN4 leucine zipper, a two-stranded, parallel coiled coil. Science. 1991;254:539–544. - PubMed
-
- Burkhard P, Stetefeld J, Strelkov SV. Coiled coils: a highly versatile protein folding motif. Trends Cell Biol. 2001;11:82–88. - PubMed
-
- Brown JH, Cohen C. Regulation of muscle contraction by tropomyosin and troponhow structure illuminates function. In: Squire JM, Parry DA, editors. Fibrous proteins and related structures. Elsevier/Academic press; 2005.
-
- Crick FHC. The packing of alpha-helices: simple coiled coils. Acta Crystallogr. 1953;6:689–697.
-
- Cohen C, Parry DAD. Alpha-helical coiled coils and bundles: how to design an alpha-helical protein. Protein Struct Funct Genet. 1990;7:1–15. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

