A computational study of RNA binding and specificity in the tandem zinc finger domain of TIS11d
- PMID: 20506496
- PMCID: PMC2895246
- DOI: 10.1002/pro.401
A computational study of RNA binding and specificity in the tandem zinc finger domain of TIS11d
Abstract
TIS11d is a member of the CCCH-type family of tandem zinc finger (TZF) proteins; the TZF domain of TIS11d (residues 151-220) is sufficient to bind and destabilize its target mRNAs with high specificity. In this study, the TZF domain of TIS11d is simulated in an aqueous environment in both the free and RNA-bound states. Multiple nanosecond timescale molecular dynamics trajectories of TIS11d wild-type and E157R/E195K mutant with different RNA sequences were performed to investigate the molecular basis for RNA binding specificities of this TZF domain. A variety of measures of the protein structure, fluctuations, and dynamics were used to analyze the trajectories. The results of this study support the following conclusions: (1) the structure of the two fingers is maintained in the free state but a global reorientation occurs to yield a more compact structure; (2) mutation of the glutamate residues at positions 157 and 195 to arginine and lysine, respectively, affects the RNA recognition by this TIS11d mutant in agreement with the findings of Pagano et al. (J Biol Chem 2007; 282:8883-8894); and (3) we predict that the E157R/E195K mutant will present a more relaxed RNA binding specificity relative to wild-type TIS11d based on the more favorable nonsequence-specific Coulomb interaction of the two positively charged residues at positions 157 and 195 with the RNA backbone, which compensates for a partial loss of the stacking interaction of aromatic side chains with the RNA bases.
Figures



 −
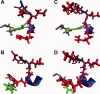
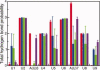
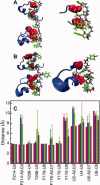
−  ; B:
; B:  −
−  .
.

References
-
- Ogilvie RL, Abelson M, Hau HH, Vlasova I, Blackshear PJ, Bohjanen PR. Tristetraprolin down-regulates IL-2 gene expression through AU-rich element-mediated mRNA decay. J Immunol. 2005;174:953–961. - PubMed
-
- Carballo E, Lai WS, Blackshear PJ. Feedback inhibition of macrophage tumor necrosis factor-alpha production by tristetraprolin. Science. 1998;281:1001–1005. - PubMed
-
- Chen CY, Gherzi R, Ong SE, Chan EL, Raijmakers R, Pruijn GJ, Stoecklin G, Moroni C, Mann M, Karin M. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell. 2001;107:451–464. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

