The cutting edge: membrane-anchored serine protease activities in the pericellular microenvironment
- PMID: 20507279
- PMCID: PMC3680374
- DOI: 10.1042/BJ20100046
The cutting edge: membrane-anchored serine protease activities in the pericellular microenvironment
Abstract
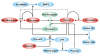
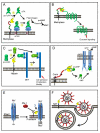
The serine proteases of the trypsin-like (S1) family play critical roles in many key biological processes including digestion, blood coagulation, and immunity. Members of this family contain N- or C-terminal domains that serve to tether the serine protease catalytic domain directly to the plasma membrane. These membrane-anchored serine proteases are proving to be key components of the cell machinery for activation of precursor molecules in the pericellular microenvironment, playing vital functions in the maintenance of homoeostasis. Substrates activated by membrane-anchored serine proteases include peptide hormones, growth and differentiation factors, receptors, enzymes, adhesion molecules and viral coat proteins. In addition, new insights into our understanding of the physiological functions of these proteases and their involvement in human pathology have come from animal models and patient studies. The present review discusses emerging evidence for the diversity of this fascinating group of membrane serine proteases as potent modifiers of the pericellular microenvironment through proteolytic processing of diverse substrates. We also discuss the functional consequences of the activities of these proteases on mammalian physiology and disease.
Figures

References
-
- Ramsay AJ, Quesada V, Sanchez M, Garabaya C, Sarda MP, Baiget M, Remacha A, Velasco G, Lopez-Otin C. Matriptase-2 mutations in iron-refractory iron deficiency anemia patients provide new insights into protease activation mechanisms. Hum. Mol. Genet. 2009;18:3673–3683. - PubMed
-
- Hedstrom L. Serine protease mechanism and specificity. Chem. Rev. 2002;102:4501–4524. - PubMed
-
- Rawlings ND, Barrett AJ. Introduction: serine peptidases and their clans. In: Barrett AJ, Rawlings ND, Woessner JF, editors. In Handbook of Proteolytic Enzymes. 2nd Edition Vol. 2. Elsevier Ltd; London UK: 2004. pp. 1417–1439.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

