Comparative genomic analysis reveals evidence of two novel Vibrio species closely related to V. cholerae
- PMID: 20507608
- PMCID: PMC2889950
- DOI: 10.1186/1471-2180-10-154
Comparative genomic analysis reveals evidence of two novel Vibrio species closely related to V. cholerae
Abstract
Background: In recent years genome sequencing has been used to characterize new bacterial species, a method of analysis available as a result of improved methodology and reduced cost. Included in a constantly expanding list of Vibrio species are several that have been reclassified as novel members of the Vibrionaceae. The description of two putative new Vibrio species, Vibrio sp. RC341 and Vibrio sp. RC586 for which we propose the names V. metecus and V. parilis, respectively, previously characterized as non-toxigenic environmental variants of V. cholerae is presented in this study.
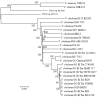
Results: Based on results of whole-genome average nucleotide identity (ANI), average amino acid identity (AAI), rpoB similarity, MLSA, and phylogenetic analysis, the new species are concluded to be phylogenetically closely related to V. cholerae and V. mimicus. Vibrio sp. RC341 and Vibrio sp. RC586 demonstrate features characteristic of V. cholerae and V. mimicus, respectively, on differential and selective media, but their genomes show a 12 to 15% divergence (88 to 85% ANI and 92 to 91% AAI) compared to the sequences of V. cholerae and V. mimicus genomes (ANI <95% and AAI <96% indicative of separate species). Vibrio sp. RC341 and Vibrio sp. RC586 share 2104 ORFs (59%) and 2058 ORFs (56%) with the published core genome of V. cholerae and 2956 (82%) and 3048 ORFs (84%) with V. mimicus MB-451, respectively. The novel species share 2926 ORFs with each other (81% Vibrio sp. RC341 and 81% Vibrio sp. RC586). Virulence-associated factors and genomic islands of V. cholerae and V. mimicus, including VSP-I and II, were found in these environmental Vibrio spp.
Conclusions: Results of this analysis demonstrate these two environmental vibrios, previously characterized as variant V. cholerae strains, are new species which have evolved from ancestral lineages of the V. cholerae and V. mimicus clade. The presence of conserved integration loci for genomic islands as well as evidence of horizontal gene transfer between these two new species, V. cholerae, and V. mimicus suggests genomic islands and virulence factors are transferred between these species.
Figures


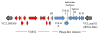
Similar articles
-
Novel Cholera Toxin Variant and ToxT Regulon in Environmental Vibrio mimicus Isolates: Potential Resources for the Evolution of Vibrio cholerae Hybrid Strains.Appl Environ Microbiol. 2019 Jan 23;85(3):e01977-18. doi: 10.1128/AEM.01977-18. Print 2019 Feb 1. Appl Environ Microbiol. 2019. PMID: 30446560 Free PMC article.
-
Genomic taxonomy of Vibrios.BMC Evol Biol. 2009 Oct 27;9:258. doi: 10.1186/1471-2148-9-258. BMC Evol Biol. 2009. PMID: 19860885 Free PMC article.
-
Genome sequencing reveals unique mutations in characteristic metabolic pathways and the transfer of virulence genes between V. mimicus and V. cholerae.PLoS One. 2011;6(6):e21299. doi: 10.1371/journal.pone.0021299. Epub 2011 Jun 22. PLoS One. 2011. PMID: 21731695 Free PMC article.
-
Biodiversity of vibrios.Microbiol Mol Biol Rev. 2004 Sep;68(3):403-31, table of contents. doi: 10.1128/MMBR.68.3.403-431.2004. Microbiol Mol Biol Rev. 2004. PMID: 15353563 Free PMC article. Review.
-
What Whole Genome Sequencing Has Told Us About Pathogenic Vibrios.Adv Exp Med Biol. 2023;1404:337-352. doi: 10.1007/978-3-031-22997-8_16. Adv Exp Med Biol. 2023. PMID: 36792883 Review.
Cited by
-
Updating the Vibrio clades defined by multilocus sequence phylogeny: proposal of eight new clades, and the description of Vibrio tritonius sp. nov.Front Microbiol. 2013 Dec 27;4:414. doi: 10.3389/fmicb.2013.00414. eCollection 2013. Front Microbiol. 2013. PMID: 24409173 Free PMC article.
-
Ethanolamine utilization in Vibrio alginolyticus.Biol Direct. 2012 Dec 12;7:45; discussion 45. doi: 10.1186/1745-6150-7-45. Biol Direct. 2012. PMID: 23234435 Free PMC article.
-
Assay for Evaluating the Abundance of Vibrio cholerae and Its O1 Serogroup Subpopulation from Water without DNA Extraction.Pathogens. 2022 Mar 16;11(3):363. doi: 10.3390/pathogens11030363. Pathogens. 2022. PMID: 35335687 Free PMC article.
-
The Vibrio cholerae fatty acid regulatory protein, FadR, represses transcription of plsB, the gene encoding the first enzyme of membrane phospholipid biosynthesis.Mol Microbiol. 2011 Aug;81(4):1020-33. doi: 10.1111/j.1365-2958.2011.07748.x. Epub 2011 Jul 19. Mol Microbiol. 2011. PMID: 21771112 Free PMC article.
-
Genetic relationships of Vibrio parahaemolyticus isolates from clinical, human carrier, and environmental sources in Thailand, determined by multilocus sequence analysis.Appl Environ Microbiol. 2013 Apr;79(7):2358-70. doi: 10.1128/AEM.03067-12. Epub 2013 Feb 1. Appl Environ Microbiol. 2013. PMID: 23377932 Free PMC article.
References
-
- Kushmaro A, Banin E, Loya Y, Stackebrandt E, Rosenberg E. Vibrio shiloi sp. nov., the causative agent of bleaching of the coral Oculina patagonica. Int J Syst Evol Microbiol. 2001;51:1383–1388. - PubMed
-
- Guerinot ML, West PA, Lee JV, Colwell RR. Vibrio diazotrophicus sp. nov., a marine nitrogen-fixing bacterium. International Journal of Systematic and Evolutionary Microbiology. 1982;32(3):350–357.
-
- Hada HS, West PA, Lee JV, Stemmler J, Colwell RR. Vibrio tubiashii sp. nov., a pathogen of bivalve mollusks. International Journal of Systematic and Evolutionary Microbiology. 1984;34(1):1–4.
-
- Hedlund BP, Staley JT. Vibrio cyclotrophicus sp. nov., a polycyclic aromatic hydrocarbon (PAH)-degrading marine bacterium. Int J Syst Evol Microbiol. 2001;51:61–66. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

