Molecular dynamics simulations suggest that RNA three-way junctions can act as flexible RNA structural elements in the ribosome
- PMID: 20507916
- PMCID: PMC2952862
- DOI: 10.1093/nar/gkq414
Molecular dynamics simulations suggest that RNA three-way junctions can act as flexible RNA structural elements in the ribosome
Abstract
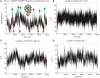
We present extensive explicit solvent molecular dynamics analysis of three RNA three-way junctions (3WJs) from the large ribosomal subunit: the 3WJ formed by Helices 90-92 (H90-H92) of 23S rRNA; the 3WJ formed by H42-H44 organizing the GTPase associated center (GAC) of 23S rRNA; and the 3WJ of 5S rRNA. H92 near the peptidyl transferase center binds the 3'-CCA end of amino-acylated tRNA. The GAC binds protein factors and stimulates GTP hydrolysis driving protein synthesis. The 5S rRNA binds the central protuberance and A-site finger (ASF) involved in bridges with the 30S subunit. The simulations reveal that all three 3WJs possess significant anisotropic hinge-like flexibility between their stacked stems and dynamics within the compact regions of their adjacent stems. The A-site 3WJ dynamics may facilitate accommodation of tRNA, while the 5S 3WJ flexibility appears to be essential for coordinated movements of ASF and 5S rRNA. The GAC 3WJ may support large-scale dynamics of the L7/L12-stalk region. The simulations reveal that H42-H44 rRNA segments are not fully relaxed and in the X-ray structures they are bent towards the large subunit. The bending may be related to L10 binding and is distributed between the 3WJ and the H42-H97 contact.
Figures
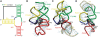



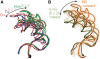

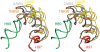
Similar articles
-
The 3D arrangement of the 23 S and 5 S rRNA in the Escherichia coli 50 S ribosomal subunit based on a cryo-electron microscopic reconstruction at 7.5 A resolution.J Mol Biol. 2000 Apr 21;298(1):35-59. doi: 10.1006/jmbi.2000.3635. J Mol Biol. 2000. PMID: 10756104
-
Elastic properties of ribosomal RNA building blocks: molecular dynamics of the GTPase-associated center rRNA.Nucleic Acids Res. 2007;35(12):4007-17. doi: 10.1093/nar/gkm245. Epub 2007 Jun 6. Nucleic Acids Res. 2007. PMID: 17553840 Free PMC article.
-
RNA tertiary interactions in the large ribosomal subunit: the A-minor motif.Proc Natl Acad Sci U S A. 2001 Apr 24;98(9):4899-903. doi: 10.1073/pnas.081082398. Epub 2001 Apr 10. Proc Natl Acad Sci U S A. 2001. PMID: 11296253 Free PMC article.
-
Structure of a two-domain N-terminal fragment of ribosomal protein L10 from Methanococcus jannaschii reveals a specific piece of the archaeal ribosomal stalk.J Mol Biol. 2010 Jun 4;399(2):214-20. doi: 10.1016/j.jmb.2010.04.017. Epub 2010 Apr 24. J Mol Biol. 2010. PMID: 20399793 Review.
-
Structure and function of ribosomal RNA.Biochem Cell Biol. 1995 Nov-Dec;73(11-12):997-1009. doi: 10.1139/o95-107. Biochem Cell Biol. 1995. PMID: 8722015 Review.
Cited by
-
Structure and mechanical properties of the ribosomal L1 stalk three-way junction.Nucleic Acids Res. 2012 Jul;40(13):6290-303. doi: 10.1093/nar/gks258. Epub 2012 Mar 26. Nucleic Acids Res. 2012. PMID: 22451682 Free PMC article.
-
Hierarchy of RNA functional dynamics.Annu Rev Biochem. 2014;83:441-66. doi: 10.1146/annurev-biochem-060713-035524. Epub 2014 Mar 5. Annu Rev Biochem. 2014. PMID: 24606137 Free PMC article. Review.
-
Platinum-RNA modifications following drug treatment in S. cerevisiae identified by click chemistry and enzymatic mapping.ACS Chem Biol. 2014 Oct 17;9(10):2404-11. doi: 10.1021/cb500395z. Epub 2014 Aug 15. ACS Chem Biol. 2014. PMID: 25055168 Free PMC article.
-
RNA as a stable polymer to build controllable and defined nanostructures for material and biomedical applications.Nano Today. 2015 Oct 1;10(5):631-655. doi: 10.1016/j.nantod.2015.09.003. Nano Today. 2015. PMID: 26770259 Free PMC article.
-
Connecting energy landscapes with experimental rates for aminoacyl-tRNA accommodation in the ribosome.J Am Chem Soc. 2010 Sep 29;132(38):13170-1. doi: 10.1021/ja1061399. J Am Chem Soc. 2010. PMID: 20806913 Free PMC article.

