Comparison of macrophage antimicrobial responses induced by type II interferons of the goldfish (Carassius auratus L.)
- PMID: 20507977
- PMCID: PMC2911290
- DOI: 10.1074/jbc.M109.096925
Comparison of macrophage antimicrobial responses induced by type II interferons of the goldfish (Carassius auratus L.)
Abstract
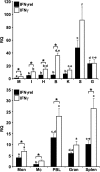
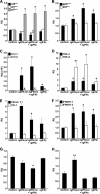
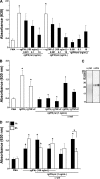
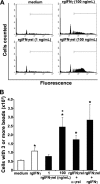
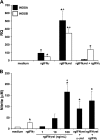
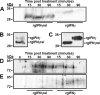
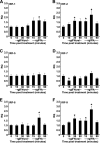
Unlike mammals, bony fish have two type II interferons, IFNgamma and IFNgammarel, whose pro-inflammatory functions have not been fully characterized. To elucidate the distinct roles of these type II interferons of bony fish, we examined the effects of recombinant goldfish (rg) IFNgamma and IFNgammarel on the macrophage antimicrobial responses, immune gene expression, and their signaling pathways. Our findings indicate that rgIFNgamma and rgIFNgammarel possess unique capacities to mediate each of the above processes. Q-PCR analysis revealed similar expression of both cytokines in tissues and immune cell populations of the goldfish, although IFNgamma mRNA levels were generally higher in most tissues and cell types. Whereas rgIFNgamma had long-lasting effects on the priming of goldfish monocyte ROI production, the rgIFNgammarel had relatively short-lived ROI priming potential and eventually down-regulated the priming of ROI production induced by rgIFNgamma or rgTNFalpha2. Whereas rgIFNgamma induced relatively modest phagocytic and nitric oxide responses of goldfish macrophages, rgIFNgammarel induced significantly higher phagocytosis, iNOSA and iNOSB gene expression and nitric oxide production compared with rgIFNgamma. The rgIFNgamma and rgIFNgammarel induced different gene expression profiles in goldfish monocytes. These differences included significantly higher induction of TNFalpha2, CXCL8, ceruloplasmin, and interferon regulatory factor (IRFs) expression after activation of monocytes with rgIFNgammarel. The rgIFNgammarel was more abundant in whole cell lysates compared with rgIFNgamma. Both cytokines induced the phosphorylation of Stat1, while the nuclear localization of Stat1 was only observed following treatment of monocytes with rgIFNgamma. Our findings suggest the presence of functional segregation of the induction of macrophage antimicrobial functions by type II interferons of bony fish.
Figures
References
-
- Mosmann T. R., Coffman R. L. (1989) Annu. Rev. Immunol. 7, 145–173 - PubMed
-
- Sad S., Marcotte R., Mosmann T. R. (1995) Immunity 2, 271–279 - PubMed
-
- Perussia B. (1991) Curr. Opin. Immunol. 3, 49–55 - PubMed
-
- Wheelock E. F. (1965) Science 149, 310–311 - PubMed
-
- Staeheli P. (1990) Adv. Virus. Res. 38, 147–200 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

