Foot-and-mouth disease virus 2C is a hexameric AAA+ protein with a coordinated ATP hydrolysis mechanism
- PMID: 20507978
- PMCID: PMC2915670
- DOI: 10.1074/jbc.M110.129940
Foot-and-mouth disease virus 2C is a hexameric AAA+ protein with a coordinated ATP hydrolysis mechanism
Abstract
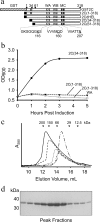
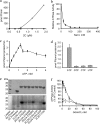
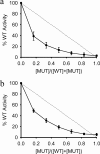
Foot-and-mouth disease virus (FMDV), a positive sense, single-stranded RNA virus, causes a highly contagious disease in cloven-hoofed livestock. Like other picornaviruses, FMDV has a conserved 2C protein assigned to the superfamily 3 helicases a group of AAA+ ATPases that has a predicted N-terminal membrane-binding amphipathic helix attached to the main ATPase domain. In infected cells, 2C is involved in the formation of membrane vesicles, where it co-localizes with viral RNA replication complexes, but its precise role in virus replication has not been elucidated. We show here that deletion of the predicted N-terminal amphipathic helix enables overexpression in Escherichia coli of a highly soluble truncated protein, 2C(34-318), that has ATPase and RNA binding activity. ATPase activity was abrogated by point mutations in the Walker A (K116A) and B (D160A) motifs and Motif C (N207A) in the active site. Unliganded 2C(34-318) exhibits concentration-dependent self-association to yield oligomeric forms, the largest of which is tetrameric. Strikingly, in the presence of ATP and RNA, FMDV 2C(34-318) containing the N207A mutation, which binds but does not hydrolyze ATP, was found to oligomerize specifically into hexamers. Visualization of FMDV 2C-ATP-RNA complexes by negative stain electron microscopy revealed hexameric ring structures with 6-fold symmetry that are characteristic of AAA+ ATPases. ATPase assays performed by mixing purified active and inactive 2C(34-318) subunits revealed a coordinated mechanism of ATP hydrolysis. Our results provide new insights into the structure and mechanism of picornavirus 2C proteins that will facilitate new investigations of their roles in infection.
Figures



Similar articles
-
Heat Shock Protein 60 Is Involved in Viral Replication Complex Formation and Facilitates Foot and Mouth Virus Replication by Stabilizing Viral Nonstructural Proteins 3A and 2C.mBio. 2022 Oct 26;13(5):e0143422. doi: 10.1128/mbio.01434-22. Epub 2022 Sep 15. mBio. 2022. PMID: 36106732 Free PMC article.
-
Poliovirus 2C protein forms homo-oligomeric structures required for ATPase activity.J Biol Chem. 2009 Aug 14;284(33):22012-22021. doi: 10.1074/jbc.M109.031807. Epub 2009 Jun 11. J Biol Chem. 2009. PMID: 19520852 Free PMC article.
-
A cardioviral 2C-ATP complex structure reveals the essential role of a conserved arginine in regulation of cardioviral 2C activity.J Virol. 2024 Oct 22;98(10):e0091124. doi: 10.1128/jvi.00911-24. Epub 2024 Sep 6. J Virol. 2024. PMID: 39240112 Free PMC article.
-
Picornavirus non-structural proteins as targets for new anti-virals with broad activity.Antiviral Res. 2011 Mar;89(3):204-18. doi: 10.1016/j.antiviral.2010.12.007. Epub 2011 Jan 12. Antiviral Res. 2011. PMID: 21236302 Review.
-
Mechanism of RNA packaging motor.Adv Exp Med Biol. 2012;726:609-29. doi: 10.1007/978-1-4614-0980-9_27. Adv Exp Med Biol. 2012. PMID: 22297533 Review.
Cited by
-
Peptides Interfering 3A Protein Dimerization Decrease FMDV Multiplication.PLoS One. 2015 Oct 27;10(10):e0141415. doi: 10.1371/journal.pone.0141415. eCollection 2015. PLoS One. 2015. PMID: 26505190 Free PMC article.
-
Discovery of Small Molecules Against Foot-and-Mouth Disease Virus Replication by Targeting 2C Helicase Activity.Viruses. 2025 May 29;17(6):785. doi: 10.3390/v17060785. Viruses. 2025. PMID: 40573376 Free PMC article.
-
The nonstructural protein 2C of a Picorna-like virus displays nucleic acid helix destabilizing activity that can be functionally separated from its ATPase activity.J Virol. 2013 May;87(9):5205-18. doi: 10.1128/JVI.00245-13. Epub 2013 Feb 28. J Virol. 2013. PMID: 23449794 Free PMC article.
-
Non-lytic spread of poliovirus requires the nonstructural protein 3CD.bioRxiv [Preprint]. 2024 Oct 19:2024.10.18.619132. doi: 10.1101/2024.10.18.619132. bioRxiv. 2024. Update in: mBio. 2025 Jan 8;16(1):e0327624. doi: 10.1128/mbio.03276-24. PMID: 39464037 Free PMC article. Updated. Preprint.
-
Heat Shock Protein 60 Is Involved in Viral Replication Complex Formation and Facilitates Foot and Mouth Virus Replication by Stabilizing Viral Nonstructural Proteins 3A and 2C.mBio. 2022 Oct 26;13(5):e0143422. doi: 10.1128/mbio.01434-22. Epub 2022 Sep 15. mBio. 2022. PMID: 36106732 Free PMC article.

