GANP-mediated recruitment of activation-induced cytidine deaminase to cell nuclei and to immunoglobulin variable region DNA
- PMID: 20507984
- PMCID: PMC2911284
- DOI: 10.1074/jbc.M110.131441
GANP-mediated recruitment of activation-induced cytidine deaminase to cell nuclei and to immunoglobulin variable region DNA
Abstract
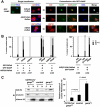
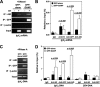
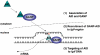
AID (activation-induced cytidine deaminase) catalyzes transcription-dependent deamination of C --> U in immunoglobulin variable (IgV) regions to initiate somatic hypermutation (SHM) in germinal center B-cells. SHM is essential in generating high affinity antibodies. Here we show that when coexpressed with GANP (germinal center-associated nuclear protein) in COS-7 cells, AID is transported from the cytoplasm and concentrated in the nucleus. GANP forms a complex with AID in cotransfected cells in vivo and in vitro. We have isolated AID mutants that bind with reduced affinity to GANP compared with wild type AID. One of these mutants, AID (D143A) binds GANP with a 10-fold lower affinity compared with wild type AID yet retains substantial C-deamination activity in vitro. Mutant AID (D143A) remains localized predominantly in the cytoplasm when coexpressed with GANP. Exogenous expression of GANP in Ramos B-cells promotes binding of AID to IgV DNA and mRNA and increases SHM frequency. These data suggest that GANP may serve as an essential link required to transport AID to B-cell nuclei and to target AID to actively transcribed IgV regions.
Figures



Similar articles
-
Integrity of immunoglobulin variable regions is supported by GANP during AID-induced somatic hypermutation in germinal center B cells.Int Immunol. 2017 May 1;29(5):211-220. doi: 10.1093/intimm/dxx032. Int Immunol. 2017. PMID: 28541550 Free PMC article.
-
GANP regulates recruitment of AID to immunoglobulin variable regions by modulating transcription and nucleosome occupancy.Nat Commun. 2013;4:1830. doi: 10.1038/ncomms2823. Nat Commun. 2013. PMID: 23652018 Free PMC article.
-
Molecular mechanism of immunoglobulin V-region diversification regulated by transcription and RNA metabolism in antigen-driven B cells.Scand J Immunol. 2011 Jun;73(6):520-6. doi: 10.1111/j.1365-3083.2011.02557.x. Scand J Immunol. 2011. PMID: 21388430 Review.
-
GANP regulates the choice of DNA repair pathway by DNA-PKcs interaction in AID-dependent IgV region diversification.J Immunol. 2014 Jun 15;192(12):5529-39. doi: 10.4049/jimmunol.1400021. Epub 2014 May 7. J Immunol. 2014. PMID: 24808370
-
Germinal Center B-Cell-Associated Nuclear Protein (GANP) Involved in RNA Metabolism for B Cell Maturation.Adv Immunol. 2016;131:135-86. doi: 10.1016/bs.ai.2016.02.003. Epub 2016 Mar 29. Adv Immunol. 2016. PMID: 27235683 Review.
Cited by
-
Activation-induced cytidine deaminase (AID)-dependent somatic hypermutation requires a splice isoform of the serine/arginine-rich (SR) protein SRSF1.Proc Natl Acad Sci U S A. 2012 Jan 24;109(4):1216-21. doi: 10.1073/pnas.1120368109. Epub 2012 Jan 9. Proc Natl Acad Sci U S A. 2012. PMID: 22232677 Free PMC article.
-
Activation-induced cytidine deaminase targets SUV4-20-mediated histone H4K20 trimethylation to class-switch recombination sites.Sci Rep. 2017 Aug 8;7(1):7594. doi: 10.1038/s41598-017-07380-9. Sci Rep. 2017. PMID: 28790320 Free PMC article.
-
Complex regulation and function of activation-induced cytidine deaminase.Trends Immunol. 2011 May;32(5):194-201. doi: 10.1016/j.it.2011.03.003. Epub 2011 Apr 13. Trends Immunol. 2011. PMID: 21493144 Free PMC article. Review.
-
A role for the RNA pol II-associated PAF complex in AID-induced immune diversification.J Exp Med. 2012 Oct 22;209(11):2099-111. doi: 10.1084/jem.20112145. Epub 2012 Sep 24. J Exp Med. 2012. PMID: 23008333 Free PMC article.
-
Intrinsic restriction activity by apolipoprotein B mRNA editing enzyme APOBEC1 against the mobility of autonomous retrotransposons.Nucleic Acids Res. 2011 Jul;39(13):5538-54. doi: 10.1093/nar/gkr124. Epub 2011 Mar 12. Nucleic Acids Res. 2011. PMID: 21398638 Free PMC article.
References
-
- MacLennan I. C. (1994) Annu. Rev. Immunol. 12, 117–139 - PubMed
-
- Rajewsky K. (1996) Nature 381, 751–758 - PubMed
-
- Muramatsu M., Kinoshita K., Fagarasan S., Yamada S., Shinkai Y., Honjo T. (2000) Cell 102, 553–563 - PubMed
-
- Di Noia J. M., Neuberger M. S. (2007) Annu. Rev. Biochem. 76, 1–22 - PubMed
-
- Peled J. U., Kuang F. L., Iglesias-Ussel M. D., Roa S., Kalis S. L., Goodman M. F., Scharff M. D. (2008) Annu. Rev. Immunol. 26, 481–511 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

