Genome-wide kinetics of nucleosome turnover determined by metabolic labeling of histones
- PMID: 20508129
- PMCID: PMC2879085
- DOI: 10.1126/science.1186777
Genome-wide kinetics of nucleosome turnover determined by metabolic labeling of histones
Abstract
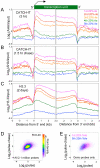
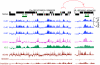
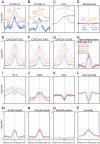
Nucleosome disruption and replacement are crucial activities that maintain epigenomes, but these highly dynamic processes have been difficult to study. Here, we describe a direct method for measuring nucleosome turnover dynamics genome-wide. We found that nucleosome turnover is most rapid over active gene bodies, epigenetic regulatory elements, and replication origins in Drosophila cells. Nucleosomes turn over faster at sites for trithorax-group than polycomb-group protein binding, suggesting that nucleosome turnover differences underlie their opposing activities and challenging models for epigenetic inheritance that rely on stability of histone marks. Our results establish a general strategy for studying nucleosome dynamics and uncover nucleosome turnover differences across the genome that are likely to have functional importance for epigenome maintenance, gene regulation, and control of DNA replication.
Figures
Comment in
-
Epigenomics: Catching nucleosomes in action.Nat Rev Genet. 2010 Jul;11(7):457. doi: 10.1038/nrg2822. Nat Rev Genet. 2010. PMID: 20531368 No abstract available.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

