Activation of plant immune responses by a gain-of-function mutation in an atypical receptor-like kinase
- PMID: 20508139
- PMCID: PMC2923897
- DOI: 10.1104/pp.110.158501
Activation of plant immune responses by a gain-of-function mutation in an atypical receptor-like kinase
Abstract
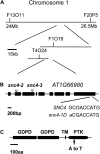
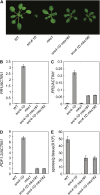
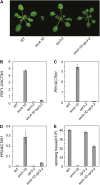
Arabidopsis (Arabidopsis thaliana) suppressor of npr1-1, constitutive1 (snc1) contains a gain-of-function mutation in a Toll/interleukin receptor-nucleotide binding site-leucine-rich repeat Resistance (R) protein and it has been a useful tool for dissecting R-protein-mediated immunity. Here we report the identification and characterization of snc4-1D, a semidominant mutant with snc1-like phenotypes. snc4-1D constitutively expresses defense marker genes PR1, PR2, and PDF1.2, and displays enhanced pathogen resistance. Map-based cloning of SNC4 revealed that it encodes an atypical receptor-like kinase with two predicted extracellular glycerophosphoryl diester phosphodiesterase domains. The snc4-1D mutation changes an alanine to threonine in the predicted cytoplasmic kinase domain. Wild-type plants transformed with the mutant snc4-1D gene displayed similar phenotypes as snc4-1D, suggesting that the mutation is a gain-of-function mutation. Epistasis analysis showed that NON-RACE-SPECIFIC DISEASE RESISTANCE1 is required for the snc4-1D mutant phenotypes. In addition, the snc4-1D mutant phenotypes are partially suppressed by knocking out MAP KINASE SUBSTRATE1, a positive defense regulator associated with MAP KINASE4. Furthermore, both the morphology and constitutive pathogen resistance of snc4-1D are partially suppressed by blocking jasmonic acid synthesis, suggesting that jasmonic acid plays an important role in snc4-1D-mediated resistance. Identification of snc4-1D provides us a unique genetic system for analyzing the signal transduction pathways downstream of receptor-like kinases.
Figures




References
-
- Boller T, Felix G. (2009) A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol 60: 379–406 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

