Nucleosome occupancy landscape and dynamics at mouse recombination hotspots
- PMID: 20508641
- PMCID: PMC2897116
- DOI: 10.1038/embor.2010.79
Nucleosome occupancy landscape and dynamics at mouse recombination hotspots
Abstract
During meiosis, paternal and maternal homologous chromosomes recombine at specific recombination sites named hotspots. What renders 2% of the mammalian genomes permissive to meiotic recombination by allowing Spo11 endonuclease to initiate double-strand breaks is largely unknown. Work in yeast has shown that chromatin accessibility seems to be important for this activity. Here, we define nucleosome profiles and dynamics at four mouse recombination hotspots by purifying highly enriched fractions of meiotic cells. We found that nucleosome occupancy is generally stable during meiosis progression. Interestingly, the cores of recombination hotspots have largely open chromatin structure, and the localization of the few nucleosomes present in these cores correlates precisely with the crossover-free zones in recombinogenic domains. Collectively, these high-resolution studies suggest that nucleosome occupancy seems to direct, at least in part, how meiotic recombination events are processed.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
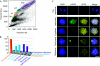

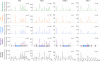
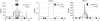
References
-
- Bastos H, Lassalle B, Chicheportiche A, Riou L, Testart J, Allemand I, Fouchet P (2005) Flow cytometric characterization of viable meiotic and postmeiotic cells by Hoechst 33342 in mouse spermatogenesis. Cytometry A 65: 40–49 - PubMed
-
- Di Felice F, Chiani F, Camilloni G (2008) Nucleosomes represent a physical barrier for cleavage activity of DNA topoisomerase I in vivo. Biochem J 409: 651–656 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

