Double displacement: An improved bioorthogonal reaction strategy for templated nucleic acid detection
- PMID: 20509625
- PMCID: PMC2891565
- DOI: 10.1021/bc100165h
Double displacement: An improved bioorthogonal reaction strategy for templated nucleic acid detection
Abstract
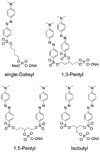
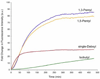
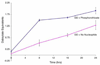
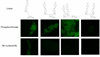
Quenched autoligation probes have been employed previously in a target-templated nonenzymatic ligation strategy for detecting nucleic acids in cells by fluorescence. A common source of background signal in such probes is the undesired reaction with water and other cellular nucleophiles. Here, we describe a new class of self-ligating probes, double displacement (DD) probes, that rely on two displacement reactions to fully unquench a nearby fluorophore. Three potential double displacement architectures, all possessing two fluorescence quencher/leaving groups (dabsylate groups), were synthesized and evaluated for templated reaction with nucleophile (phosphorothioate) probes both in vitro and in intact bacterial cells. All three DD probe designs provided substantially better initial quenching than a single-Dabsyl control. In isothermal templated reactions in vitro, double displacement probes yielded considerably lower background signal than previous single displacement probes; investigation into the mechanism revealed that one dabsylate acts as a sacrificial leaving group, reacting nonspecifically with water, but yielding little signal because another quencher group remains. Templated reaction with the specific nucleophile probe is required to activate a signal. The double displacement probes provided a ca. 80-fold turn-on signal and yielded a 2-4-fold improvement in signal/background over single Dabsyl probes. The best-performing probe architecture was demonstrated in a two-color, FRET-based two-allele discrimination system in vitro and was shown to be capable of discriminating between two closely related species of bacteria differing by a single nucleotide at an rRNA target site.
Figures


Similar articles
-
Efficient nucleic acid detection by templated reductive quencher release.J Am Chem Soc. 2009 Nov 11;131(44):16021-3. doi: 10.1021/ja904138v. J Am Chem Soc. 2009. PMID: 19886694 Free PMC article.
-
Real-time, sequence-specific detection of nucleic acids during strand displacement amplification.Anal Biochem. 1999 Dec 15;276(2):177-87. doi: 10.1006/abio.1999.4350. Anal Biochem. 1999. PMID: 10603241
-
Nonenzymatic DNA ligation in Escherichia coli cells.Nucleic Acids Res Suppl. 2002;(2):121-2. doi: 10.1093/nass/2.1.121. Nucleic Acids Res Suppl. 2002. PMID: 12903135
-
Homogeneous detection of nucleic acids using self-quenched polymerase chain reaction primers labeled with a single fluorophore (LUX primers).Methods Mol Biol. 2006;335:95-114. doi: 10.1385/1-59745-069-3:95. Methods Mol Biol. 2006. PMID: 16785623 Review.
-
Detecting RNA/DNA hybridization using double-labeled donor probes with enhanced fluorescence resonance energy transfer signals.Methods Mol Biol. 2006;335:43-56. doi: 10.1385/1-59745-069-3:43. Methods Mol Biol. 2006. PMID: 16785619 Review.
Cited by
-
Chemically modified nucleic acids and DNA intercalators as tools for nanoparticle assembly.Chem Soc Rev. 2021 Nov 29;50(23):13410-13440. doi: 10.1039/d1cs00632k. Chem Soc Rev. 2021. PMID: 34792047 Free PMC article. Review.
-
Sandwich probes: two simultaneous reactions for templated nucleic acid detection.Chem Commun (Camb). 2010 Nov 21;46(43):8154-6. doi: 10.1039/c0cc01968b. Epub 2010 Oct 7. Chem Commun (Camb). 2010. PMID: 20927470 Free PMC article.
-
Two successive reactions on a DNA template: a strategy for improving background fluorescence and specificity in nucleic acid detection.Chemistry. 2011 Feb 11;17(7):2168-75. doi: 10.1002/chem.201002426. Epub 2011 Jan 10. Chemistry. 2011. PMID: 21294182 Free PMC article.
-
Improved templated fluorogenic probes enhance the analysis of closely related pathogenic bacteria by microscopy and flow cytometry.Bioconjug Chem. 2011 Sep 21;22(9):1869-77. doi: 10.1021/bc2003567. Epub 2011 Aug 26. Bioconjug Chem. 2011. PMID: 21870777 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

