Natural products as chemical probes
- PMID: 20509672
- PMCID: PMC2926141
- DOI: 10.1021/cb100105c
Natural products as chemical probes
Abstract
Natural products have evolved to encompass a broad spectrum of chemical and functional diversity. It is this diversity, along with their structural complexity, that enables nature's small molecules to target a nearly limitless number of biological macromolecules and to often do so in a highly selective fashion. Because of these characteristics, natural products have seen great success as therapeutic agents. However, this vast pool of compounds holds much promise beyond the development of future drugs. These features also make them ideal tools for the study of biological systems. Recent examples of the use of natural products and their derivatives as chemical probes to explore biological phenomena and assemble biochemical pathways are presented here.
Figures


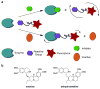

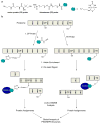
References
-
- Sertuerner F. Ueber das Morphium, eine neue salzfähige Grundlage, und die Mekonsäure, als Hauptbestandtheile des Opiums. Ann Physik. 1817;55:56–89.
-
- Newman DJ, Cragg GM. Natural Products as Drugs and Leads to Drugs: The Historical Perspective. In: Buss AD, Butler MS, editors. RSC Biomolecular Sciences No. 18; Natural Product Chemistry for Drug Discovery. Royal Society of Chemistry; Cambridge, UK: 2010. pp. 3–27.
-
- Li JWH, Vederas JC. Drug discovery and natural products: end of an era or an endless frontier? Science. 2009;325:161–165. - PubMed
-
- Newman DJ, Cragg GM, Snader KM. Natural products as sources of new drugs over the period 1981–2002. J Nat Prod. 2003;66:1022–1037. - PubMed
-
- Harvey A. Strategies for discovering drugs from previously unexplored natural products. Drug Discov Today. 2000;5:294–300. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

