Novel structural features in two ZHX homeodomains derived from a systematic study of single and multiple domains
- PMID: 20509910
- PMCID: PMC2893186
- DOI: 10.1186/1472-6807-10-13
Novel structural features in two ZHX homeodomains derived from a systematic study of single and multiple domains
Abstract
Background: Zhx1 to 3 (zinc-fingers and homeoboxes) form a set of paralogous genes encoding multi-domain proteins. ZHX proteins consist of two zinc fingers followed by five homeodomains. ZHXs have biological roles in cell cycle control by acting as co-repressors of the transcriptional regulator Nuclear Factor Y. As part of a structural genomics project we have expressed single and multi-domain fragments of the different human ZHX genes for use in structure determination.
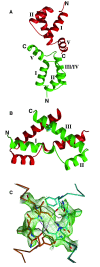
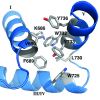
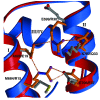
Results: A total of 30 single and multiple domain ZHX1-3 constructs selected from bioinformatics protocols were screened for soluble expression in E. coli using high throughput methodologies. Two homeodomains were crystallized leading to structures for ZHX1 HD4 and ZHX2 HD2. ZHX1 HD4, although closest matched to homeodomains from 'homez' and 'engrailed', showed structural differences, notably an additional C-terminal helix (helix V) which wrapped over helix I thereby making extensive contacts. Although ZHX2 HD2-3 was successfully expressed and purified, proteolysis occurred during crystallization yielding crystals of just HD2. The structure of ZHX2 HD2 showed an unusual open conformation with helix I undergoing 'domain-swapping' to form a homodimer.
Conclusions: Although multiple-domain constructs of ZHX1 selected by bioinformatics studies could be expressed solubly, only single homeodomains yielded crystals. The crystal structure of ZHX1 HD4 showed additional hydrophobic interactions relative to many known homeodomains via extensive contacts formed by the novel C-terminal helix V with, in particular, helix I. Additionally, the replacement of some charged covariant residues (which are commonly observed to form salt bridges in non-homeotherms such as the Drosophila 'engrailed' homeodomain), by apolar residues further increases hydrophobic contacts within ZHX1 HD4, and potentially stability, relative to engrailed homeodomain. ZHX1 HD4 helix V points away from the normally observed DNA major groove binding site on homeodomains and thus would not obstruct the putative binding of nucleic acid. In contrast, for ZHX2 HD2 the observed altered conformation involving rearrangement of helix I, relative to the canonical homeodomain fold, disrupts the normal DNA binding site, although protein-protein binding is possible as observed in homodimer formation.
Figures

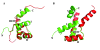
Similar articles
-
Zinc-fingers and homeoboxes (ZHX) 2, a novel member of the ZHX family, functions as a transcriptional repressor.Biochem J. 2003 Aug 1;373(Pt 3):747-57. doi: 10.1042/BJ20030171. Biochem J. 2003. PMID: 12741956 Free PMC article.
-
Analysis of zinc-fingers and homeoboxes (ZHX)-1-interacting proteins: molecular cloning and characterization of a member of the ZHX family, ZHX3.Biochem J. 2003 Jul 1;373(Pt 1):167-78. doi: 10.1042/BJ20021866. Biochem J. 2003. PMID: 12659632 Free PMC article.
-
Evolutionary Analysis of the Zinc Finger and Homeoboxes Family of Proteins Identifies Multiple Conserved Domains and a Common Early Chordate Ancestor.Genome Biol Evol. 2020 Mar 1;12(3):174-184. doi: 10.1093/gbe/evaa039. Genome Biol Evol. 2020. PMID: 32125369 Free PMC article.
-
Zinc Fingers and Homeobox Family in Cancer: A Double-Edged Sword.Int J Mol Sci. 2022 Sep 22;23(19):11167. doi: 10.3390/ijms231911167. Int J Mol Sci. 2022. PMID: 36232466 Free PMC article. Review.
-
POU domain factors in neural development.Adv Exp Med Biol. 1998;449:39-53. doi: 10.1007/978-1-4615-4871-3_4. Adv Exp Med Biol. 1998. PMID: 10026784 Review.
Cited by
-
Solution NMR structures of homeodomains from human proteins ALX4, ZHX1, and CASP8AP2 contribute to the structural coverage of the Human Cancer Protein Interaction Network.J Struct Funct Genomics. 2014 Dec;15(4):201-7. doi: 10.1007/s10969-014-9184-z. Epub 2014 Jun 19. J Struct Funct Genomics. 2014. PMID: 24941917 Free PMC article.
-
ZHX2 promotes HIF1α oncogenic signaling in triple-negative breast cancer.Elife. 2021 Nov 15;10:e70412. doi: 10.7554/eLife.70412. Elife. 2021. PMID: 34779768 Free PMC article.
-
Zinc fingers and homeoboxes 2 inhibits hepatocellular carcinoma cell proliferation and represses expression of Cyclins A and E.Gastroenterology. 2012 Jun;142(7):1559-70.e2. doi: 10.1053/j.gastro.2012.02.049. Epub 2012 Mar 6. Gastroenterology. 2012. PMID: 22406477 Free PMC article.
-
Identification of methylation-sensitive human transcription factors using meSMiLE-seq.bioRxiv [Preprint]. 2024 Nov 12:2024.11.11.619598. doi: 10.1101/2024.11.11.619598. bioRxiv. 2024. PMID: 39605503 Free PMC article. Preprint.
-
Host Transcription Factors in Hepatitis B Virus RNA Synthesis.Viruses. 2020 Jan 30;12(2):160. doi: 10.3390/v12020160. Viruses. 2020. PMID: 32019103 Free PMC article. Review.
References
-
- Yamada K, Kawata H, Shou Z, Hirano S, Mizutani T, Yazawa T, Sekiguchi T, Yoshino M, Kajitani T, Miyamoto K. Analysis of zinc-fingers and homeoboxes (ZHX)-1-interacting proteins: molecular cloning and characterization of a member of the ZHX family, ZHX3. Biochem J. 2003;373(Pt 1):167–178. doi: 10.1042/BJ20021866. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

