The vitamin D receptor: new paradigms for the regulation of gene expression by 1,25-dihydroxyvitamin D(3)
- PMID: 20511050
- PMCID: PMC2879406
- DOI: 10.1016/j.ecl.2010.02.007
The vitamin D receptor: new paradigms for the regulation of gene expression by 1,25-dihydroxyvitamin D(3)
Abstract
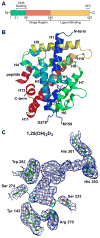
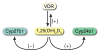
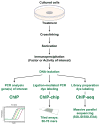
The actions of the vitamin D hormone 1,25-dihydroxyvitamin D(3) (1,25(OH)(2)D(3)) are mediated by the vitamin D receptor (VDR), a ligand-activated transcription factor that functions to control gene expression. After ligand activation, the VDR binds directly to specific sequences located near promoters and recruits a variety of coregulatory complexes that perform the additional functions required to modify transcriptional output. Recent advances in transcriptional regulation, which permit the unbiased identification of the regulatory regions of genes, are providing new insight into how genes are regulated. Surprisingly, gene regulation requires the orchestrated efforts of multiple modular enhancers often located many kilobases upstream, downstream, or within the transcription units themselves. These studies are transforming our understanding of how 1,25(OH)(2)D(3) regulates gene transcription.
Copyright 2010 Elsevier Inc. All rights reserved.
Figures
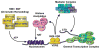
Republished in
-
The vitamin D receptor: new paradigms for the regulation of gene expression by 1,25-dihydroxyvitamin D3.Rheum Dis Clin North Am. 2012 Feb;38(1):13-27. doi: 10.1016/j.rdc.2012.03.004. Epub 2012 Apr 12. Rheum Dis Clin North Am. 2012. PMID: 22525840 Free PMC article.
References
-
- Haussler MR, Whitfield GK, Haussler CA, Hsieh JC, Thompson PD, Selznick SH, Dominguez CE, Jurutka PW. The nuclear vitamin D receptor: biological and molecular regulatory properties revealed. J Bone Miner Res. 1998 Mar;13(3):325–49. - PubMed
-
- Sutton AL, MacDonald PN. Vitamin D: more than a “bone-a-fide” hormone. Mol Endocrinol. 2003 May;17(5):777–91. - PubMed
-
- Zella LA, Kim S, Shevde NK, Pike JW. Enhancers located within two introns of the vitamin D receptor gene mediate transcriptional autoregulation by 1,25-dihydroxyvitamin D3. Mol Endocrinol. 2006 Jun;20(6):1231–47. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

