Plastidial thioredoxin z interacts with two fructokinase-like proteins in a thiol-dependent manner: evidence for an essential role in chloroplast development in Arabidopsis and Nicotiana benthamiana
- PMID: 20511297
- PMCID: PMC2899873
- DOI: 10.1105/tpc.109.071001
Plastidial thioredoxin z interacts with two fructokinase-like proteins in a thiol-dependent manner: evidence for an essential role in chloroplast development in Arabidopsis and Nicotiana benthamiana
Abstract
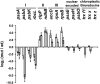
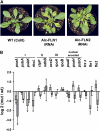
Here, we characterize a plastidial thioredoxin (TRX) isoform from Arabidopsis thaliana that defines a previously unknown branch of plastidial TRXs lying between x- and y-type TRXs and thus was named TRX z. An Arabidopsis knockout mutant of TRX z had a severe albino phenotype and was inhibited in chloroplast development. Quantitative real-time RT-PCR analysis of the mutant suggested that the expressions of genes that depend on a plastid-encoded RNA polymerase (PEP) were specifically decreased. Similar results were obtained upon virus-induced gene silencing (VIGS) of the TRX z ortholog in Nicotiana benthamiana. We found that two fructokinase-like proteins (FLN1 and FLN2), members of the pfkB-carbohydrate kinase family, were potential TRX z target proteins and identified conserved Cys residues mediating the FLN-TRX z interaction. VIGS in N. benthamiana and inducible RNA interference in Arabidopsis of FLNs also led to a repression of PEP-dependent gene transcription. Remarkably, recombinant FLNs displayed no detectable sugar-phosphorylating activity, and amino acid substitutions within the predicted active site imply that the FLNs have acquired a new function, which might be regulatory rather than metabolic. We were able to show that the FLN2 redox state changes in vivo during light/dark transitions and that this change is mediated by TRX z. Taken together, our data strongly suggest an important role for TRX z and both FLNs in the regulation of PEP-dependent transcription in chloroplasts.
Figures







Comment in
-
A new thioredoxin is involved in plastid gene expression.Plant Cell. 2010 May;22(5):1423. doi: 10.1105/tpc.110.220512. Epub 2010 May 28. Plant Cell. 2010. PMID: 20511298 Free PMC article. No abstract available.
References
-
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215: 403–410 - PubMed
-
- Balmer Y., Koller A., Val G.D., Schürmann P., Buchanan B.B. (2004a). Proteomics uncovers proteins interacting electrostatically with thioredoxin in chloroplasts. Photosynth. Res. 79: 275–280 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

