Optimizing a proteomics platform for urine biomarker discovery
- PMID: 20511394
- PMCID: PMC2957724
- DOI: 10.1074/mcp.M110.000992
Optimizing a proteomics platform for urine biomarker discovery
Abstract
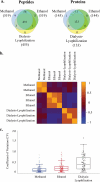
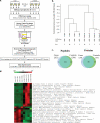
Biomarker discovery approaches in urine have been hindered by concerns for reproducibility and inadequate standardization of proteomics protocols. In this study, we describe an optimized quantitative proteomics strategy for urine biomarker discovery, which is applicable to fresh or long frozen samples. We used urine from healthy controls to standardize iTRAQ (isobaric tags for relative and absolute quantitation) for variation induced by protease inhibitors, starting protein and iTRAQ label quantities, protein extraction methods, and depletion of albumin and immunoglobulin G (IgG). We observed the following: (a) Absence of protease inhibitors did not affect the number or identity of the high confidence proteins. (b) Use of less than 20 μg of protein per sample led to a significant drop in the number of identified proteins. (c) Use of as little as a quarter unit of an iTRAQ label did not affect the number or identity of the identified proteins. (d) Protein extraction by methanol precipitation led to the highest protein yields and the most reproducible spectra. (e) Depletion of albumin and IgG did not increase the number of identified proteins or deepen the proteome coverage. Applying this optimized protocol to four pairs of long frozen urine samples from diabetic Pima Indians with or without nephropathy, we observed patterns suggesting segregation of cases and controls by iTRAQ spectra. We also identified several previously reported candidate biomarkers that showed trends toward differential expression, albeit not reaching statistical significance in this small sample set.
Figures



References
-
- Pieper R., Gatlin C. L., McGrath A. M., Makusky A. J., Mondal M., Seonarain M., Field E., Schatz C. R., Estock M. A., Ahmed N., Anderson N. G., Steiner S. (2004) Characterization of the human urinary proteome: a method for high-resolution display of urinary proteins on two-dimensional electrophoresis gels with a yield of nearly 1400 distinct protein spots. Proteomics 4, 1159–1174 - PubMed
-
- Ross P. L., Huang Y. N., Marchese J. N., Williamson B., Parker K., Hattan S., Khainovski N., Pillai S., Dey S., Daniels S., Purkayastha S., Juhasz P., Martin S., Bartlet-Jones M., He F., Jacobson A., Pappin D. J. (2004) Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell. Proteomics 3, 1154–1169 - PubMed
-
- Khan A., Packer N. H. (2006) Simple urinary sample preparation for proteomic analysis. J. Proteome Res. 5, 2824–2838 - PubMed
-
- Thongboonkerd V., Chutipongtanate S., Kanlaya R. (2006) Systematic evaluation of sample preparation methods for gel-based human urinary proteomics: quantity, quality, and variability. J. Proteome Res. 5, 183–191 - PubMed
-
- Zerefos P. G., Vlahou A. (2008) Urine sample preparation and protein profiling by two-dimensional electrophoresis and matrix-assisted laser desorption ionization time of flight mass spectroscopy. Methods Mol. Biol. 428, 141–157 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

