Replication of Vibrio cholerae chromosome I in Escherichia coli: dependence on dam methylation
- PMID: 20511501
- PMCID: PMC2916376
- DOI: 10.1128/JB.00311-10
Replication of Vibrio cholerae chromosome I in Escherichia coli: dependence on dam methylation
Abstract
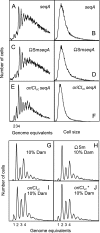
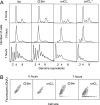
We successfully substituted Escherichia coli's origin of replication oriC with the origin region of Vibrio cholerae chromosome I (oriCI(Vc)). Replication from oriCI(Vc) initiated at a similar or slightly reduced cell mass compared to that of normal E. coli oriC. With respect to sequestration-dependent synchrony of initiation and stimulation of initiation by the loss of Hda activity, replication initiation from oriC and oriCI(Vc) were similar. Since Hda is involved in the conversion of DnaA(ATP) (DnaA bound to ATP) to DnaA(ADP) (DnaA bound to ADP), this indicates that DnaA associated with ATP is limiting for V. cholerae chromosome I replication, which similar to what is observed for E. coli. No hda homologue has been identified in V. cholerae yet. In V. cholerae, dam is essential for viability, whereas in E. coli, dam mutants are viable. Replacement of E. coli oriC with oriCI(Vc) allowed us to specifically address the role of the Dam methyltransferase and SeqA in replication initiation from oriCI(Vc). We show that when E. coli's origin of replication is substituted by oriCI(Vc), dam, but not seqA, becomes important for growth, arguing that Dam methylation exerts a critical function at the origin of replication itself. We propose that Dam methylation promotes DnaA-assisted successful duplex opening and replisome assembly at oriCI(Vc) in E. coli. In this model, methylation at oriCI(Vc) would ease DNA melting. This is supported by the fact that the requirement for dam can be alleviated by increasing negative supercoiling of the chromosome through oversupply of the DNA gyrase or loss of SeqA activity.
Figures
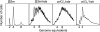


References
-
- Adamcik, J., V. Viglasky, F. Valle, M. Antalik, D. Podhradsky, and G. Dietler. 2002. Effect of bacteria growth temperature on the distribution of supercoiled DNA and its thermal stability. Electrophoresis 23:3300-3309. - PubMed
-
- Adhya, S., and M. Gottesman. 1982. Promoter occlusion: transcription through a promoter may inhibit its activity. Cell 29:939-944. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

