Three prime exonuclease I (TREX1) is Fos/AP-1 regulated by genotoxic stress and protects against ultraviolet light and benzo(a)pyrene-induced DNA damage
- PMID: 20511593
- PMCID: PMC2965218
- DOI: 10.1093/nar/gkq455
Three prime exonuclease I (TREX1) is Fos/AP-1 regulated by genotoxic stress and protects against ultraviolet light and benzo(a)pyrene-induced DNA damage
Abstract
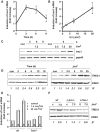
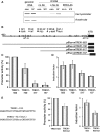
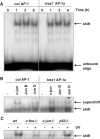
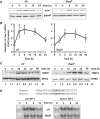
Cells respond to genotoxic stress with the induction of DNA damage defence functions. Aimed at identifying novel players in this response, we analysed the genotoxic stress-induced expression of DNA repair genes in mouse fibroblasts proficient and deficient for c-Fos or c-Jun. The experiments revealed a clear up-regulation of the three prime exonuclease I (trex1) mRNA following ultraviolet (UV) light treatment. This occurred in the wild-type but not c-fos and c-jun null cells, indicating the involvement of AP-1 in trex1 induction. Trex1 up-regulation was also observed in human cells and was found on promoter, RNA and protein level. Apart from UV light, TREX1 is induced by other DNA damaging agents such as benzo(a)pyrene and hydrogen peroxide. The mouse and human trex1 promoter harbours an AP-1 binding site that is recognized by c-Fos and c-Jun, and its mutational inactivation abrogated trex1 induction. Upon genotoxic stress, TREX1 is not only up-regulated but also translocated into the nucleus. Cells deficient in TREX1 show reduced recovery from the UV and benzo(a)pyrene-induced replication inhibition and increased sensitivity towards the genotoxins compared to the isogenic control. The data revealed trex1 as a novel DNA damage-inducible repair gene that plays a protective role in the genotoxic stress response.
Figures


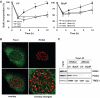
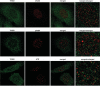
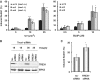
References
-
- Christmann M, Tomicic MT, Roos WP, Kaina B. Mechanisms of human DNA repair: an update. Toxicology. 2003;193:3–34. - PubMed
-
- Christmann M, Fritz G, Kaina B. Induction of DNA repair genes in mammalian cells in response to genotoxic stress. In: Lankenan D-H, editor. Genome Integrity, Facects and Perspectives. Berlin: Springer; 2007. pp. 383–398.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

