Superoxide dismutase downregulation in osteoarthritis progression and end-stage disease
- PMID: 20511611
- PMCID: PMC3789136
- DOI: 10.1136/ard.2009.119966
Superoxide dismutase downregulation in osteoarthritis progression and end-stage disease
Erratum in
- Ann Rheum Dis. 2011 Feb;70(2):397. Talyor, Robert W [corrected to Taylor, Robert W]
Abstract
Background: Oxidative stress is proposed as an important factor in osteoarthritis (OA).
Objective: To investigate the expression of the three superoxide dismutase (SOD) antioxidant enzymes in OA.
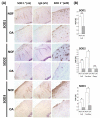
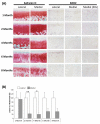
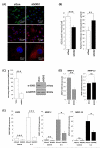
Methods: SOD expression was determined by real-time PCR and immunohistochemistry using human femoral head cartilage. SOD2 expression in Dunkin-Hartley guinea pig knee articular cartilage was determined by immunohistochemistry. The DNA methylation status of the SOD2 promoter was determined using bisulphite sequencing. RNA interference was used to determine the consequence of SOD2 depletion on the levels of reactive oxygen species (ROS) using MitoSOX and collagenases, matrix metalloproteinase 1 (MMP-1) and MMP-13, gene expression.
Results: All three SOD were abundantly expressed in human cartilage but were markedly downregulated in end-stage OA cartilage, especially SOD2. In the Dunkin-Hartley guinea pig spontaneous OA model, SOD2 expression was decreased in the medial tibial condyle cartilage before, and after, the development of OA-like lesions. The SOD2 promoter had significant DNA methylation alterations in OA cartilage. Depletion of SOD2 in chondrocytes increased ROS but decreased collagenase expression.
Conclusion: This is the first comprehensive expression profile of all SOD genes in cartilage and, importantly, using an animal model, it has been shown that a reduction in SOD2 is associated with the earliest stages of OA. A decrease in SOD2 was found to be associated with an increase in ROS but a reduction of collagenase gene expression, demonstrating the complexities of ROS function.
Figures

References
-
- Martin JA, Buckwalter JA. Aging, articular cartilage chondrocyte senescence and osteoarthritis. Biogerontology. 2002;3(5):257–64. - PubMed
-
- Carrington JL. Aging bone and cartilage: cross-cutting issues. Biochem Biophys Res Commun. 2005 Mar 18;328(3):700–8. - PubMed
-
- Henrotin Y, Kurz B, Aigner T. Oxygen and reactive oxygen species in cartilage degradation: friends or foes? Osteoarthritis Cartilage. 2005 Aug;13(8):643–54. - PubMed
-
- Plumb MS, Aspden RM. The response of elderly human articular cartilage to mechanical stimuli in vitro. Osteoarthritis Cartilage. 2005 Dec;13(12):1084–91. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

