Analysis of hairless corepressor mutants to characterize molecular cooperation with the vitamin D receptor in promoting the mammalian hair cycle
- PMID: 20512927
- PMCID: PMC2879709
- DOI: 10.1002/jcb.22578
Analysis of hairless corepressor mutants to characterize molecular cooperation with the vitamin D receptor in promoting the mammalian hair cycle
Abstract
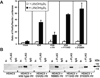
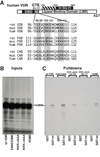
The mammalian hair cycle requires both the vitamin D receptor (VDR) and the hairless (Hr) corepressor, each of which is expressed in the hair follicle. Hr interacts directly with VDR to repress VDR-targeted transcription. Herein, we further map the VDR-interaction domain to regions in the C-terminal half of Hr that contain two LXXLL-like pairs of motifs known to mediate contact of Hr with the RAR-related orphan receptor alpha and with the thyroid hormone receptor, respectively. Site-directed mutagenesis indicates that all four hydrophobic motifs are required for VDR transrepression by Hr. Point mutation of rat Hr at conserved residues corresponding to natural mutants causing alopecia in mice (G985W and a C-terminal deletion DeltaAK) and in humans (P95S, C422Y, E611G, R640Q, C642G, N988S, D1030N, A1040T, V1074M, and V1154D), as well as alteration of residues in the C-terminal Jumonji C domain implicated in histone demethylation activity (C1025G/E1027G and H1143G) revealed that all Hr mutants retained VDR association, and that transrepressor activity was selectively abrogated in C642G, G985W, N988S, D1030N, V1074M, H1143G, and V1154D. Four of these latter Hr mutants (C642G, N988S, D1030N, and V1154D) were found to associate normally with histone deacetylase-3. Finally, we identified three regions of human VDR necessary for association with Hr, namely residues 109-111, 134-201, and 202-303. It is concluded that Hr and VDR interact via multiple protein-protein interfaces, with Hr recruiting histone deacetylases and possibly itself catalyzing histone demethylation to effect chromatin remodeling and repress the transcription of VDR target genes that control the hair cycle.
(c) 2010 Wiley-Liss, Inc.
Figures






References
-
- Ahmad W, Faiyaz ul Haque M, Brancolini V, Tsou HC, ul Haque S, Lam H, Aita VM, Owen J, deBlaquiere M, Frank J, Cserhalmi-Friedman PB, Leask A, McGrath JA, Peacocke M, Ahmad M, Ott J, Christiano AM. Alopecia universalis associated with a mutation in the human hairless gene. Science. 1998;279:720–724. - PubMed
-
- Ahmad W, Zlotogorski A, Panteleyev AA, Lam H, Ahmad M, ul Haque MF, Abdallah HM, Dragan L, Christiano AM. Genomic organization of the human hairless gene (HR) and identification of a mutation underlying congenital atrichia in an Arab Palestinian family. Genomics. 1999;56:141–148. - PubMed
-
- Aita VM, Ahmad W, Panteleyev AA, Kozlowska U, Kozlowska A, Gilliam TC, Jablonska S, Christiano AM. A novel missense mutation (C622G) in the zinc-finger domain of the human hairless gene associated with congenital atrichia with papular lesions. Exp Dermatol. 2000;9:157–162. - PubMed
-
- Bergman R, Schein-Goldshmid R, Hochberg Z, Ben-Izhak O, Sprecher E. The alopecias associated with vitamin D-dependent rickets type IIA and with hairless gene mutations: a comparative clinical, histologic, and immunohistochemical study. Arch Dermatol. 2005;141:343–351. - PubMed
-
- Brancaz MV, Iratni R, Morrison A, Mancini SJ, Marche P, Sundberg J, Nonchev S. A new allele of the mouse hairless gene interferes with Hox/LacZ transgene regulation in hair follicle primordia. Exp Mol Pathol. 2004;76:173–181. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

